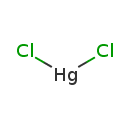
Mercuric chloride (T3D0242)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-03-06 18:58:21 UTC | |||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:21:24 UTC | |||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D0242 | |||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Mercuric chloride | |||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||
| Description | Mercuric chloride is one of the most toxic chemical compounds of mercury, due to its high solubility in water. It is most often used as a laboratory reagent, and occasionally used to form amalgams with metals. | |||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | Cl2Hg | |||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 271.500 g/mol | |||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 271.908 g/mol | |||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 7487-94-7 | |||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | dichloromercury | |||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | mercuric chloride | |||||||||||||||||||||||||||||||||||||||||||||
| SMILES | Cl[Hg]Cl | |||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/2ClH.Hg/h2*1H;/q;;+2/p-2 | |||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=LWJROJCJINYWOX-UHFFFAOYSA-L | |||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of inorganic compounds known as transition metal chlorides. These are inorganic compounds in which the largest halogen atom is Chlorine, and the heaviest metal atom is a transition metal. | |||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Inorganic compounds | |||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Mixed metal/non-metal compounds | |||||||||||||||||||||||||||||||||||||||||||||
| Class | Transition metal salts | |||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Transition metal chlorides | |||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Transition metal chlorides | |||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White crystals. | |||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Oral (8) ; inhalation (8) ; dermal (8) | |||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | High-affinity binding of the divalent mercuric ion to thiol or sulfhydryl groups of proteins is believed to be the major mechanism for the activity of mercury. Through alterations in intracellular thiol status, mercury can promote oxidative stress, lipid peroxidation, mitochondrial dysfunction, and changes in heme metabolism. Mercury is known to bind to microsomal and mitochondrial enzymes, resulting in cell injury and death. For example, mercury is known to inhibit aquaporins, halting water flow across the cell membrane. It also inhibits the protein LCK, which causes decreased T-cell signalling and immune system depression. Mercury is also believed to inhibit neuronal excitability by acting on the postsynaptic neuronal membrane. It also affects the nervous system by inhibiting protein kinase C and alkaline phosphatase, which impairs brain microvascular formation and function, as well as alters the blood-brain barrier. Mercury also produces an autoimmune response, likely by modification of major histocompatibility complex (MHC) class II molecules, self peptides, T-cell receptors, or cell-surface adhesion molecules. (8, 4, 5, 6) | |||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Mercury is absorbed mainly via ingestion and inhalation, then distributed throughout the body via the bloodstream, where a portion binds to sulfhydryl groups on haemoglobin. Mercury can undergo oxidation to mercuric mercury, which takes place via the catalase-hydrogen peroxide pathway. The mercury atom is able to diffuse down the cleft in the catalase enzyme to reach the active site where the heme ring is located. Oxidation most likely occurs in all tissue, as the catalase hydrogen peroxide pathway is ubiquitous. Following oxidation, mercury tends to accumulate in the kidneys. Mercury is excreted mainly by exhalation and in the faeces. (2, 8) | |||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 1 mg/kg (Oral, Rat) (14) LD50: 5 mg/kg (Intraperitoneal, Mouse) (14) LD50: 14 mg/kg (Subcutaneous, Rat) (14) | |||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | 1 gram for an adult human (average for inorganic mercurials). (7) | |||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | 3, not classifiable as to its carcinogenicity to humans. (12) | |||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Mercuric chloride is most often used as a laboratory reagent, and occasionally used to form an amalgam with metals. (13) | |||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Chronic Inhalation: 0.0002 mg/m3 (11) | |||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Mercury mainly affects the nervous system. Exposure to high levels of metallic, inorganic, or organic mercury can permanently damage the brain, kidneys, and developing fetus. Effects on brain functioning may result in irritability, shyness, tremors, changes in vision or hearing, and memory problems. Acrodynia, a type of mercury poisoning in children, is characterized by pain and pink discoloration of the hands and feet. Mercury poisoning can also cause Hunter-Russell syndrome and Minamata disease. (8) | |||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Common symptoms include peripheral neuropathy (presenting as paresthesia or itching, burning or pain), skin discoloration (pink cheeks, fingertips and toes), edema (swelling), and desquamation (dead skin peels off in layers). (1) | |||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Mercury poisoning is treated by immediate decontamination and chelation therapy using DMSA, DMPS, DPCN, or dimercaprol. (3) | |||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 24085 | |||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 22517 | |||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C13377 | |||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 31823 | |||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | D008627 | |||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Mercuric chloride | |||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | 826 | |||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D0242.pdf | |||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Pyrophosphatase activity
- Specific Function:
- This isozyme may play a role in skeletal mineralization.
- Gene Name:
- ALPL
- Uniprot ID:
- P05186
- Molecular Weight:
- 57304.435 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Water transmembrane transporter activity
- Specific Function:
- Forms a water-specific channel that provides the plasma membranes of red cells and kidney proximal tubules with high permeability to water, thereby permitting water to move in the direction of an osmotic gradient.
- Gene Name:
- AQP1
- Uniprot ID:
- P29972
- Molecular Weight:
- 28525.68 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Water channel required to promote glycerol permeability and water transport across cell membranes. May contribute to water transport in the upper portion of small intestine. Isoform 2 is not permeable to urea and glycerol.
- Gene Name:
- AQP10
- Uniprot ID:
- Q96PS8
- Molecular Weight:
- 31762.97 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Transporter activity
- Specific Function:
- Aquaporins facilitate the transport of water and small neutral solutes across cell membranes.
- Gene Name:
- AQP11
- Uniprot ID:
- Q8NBQ7
- Molecular Weight:
- 30202.59 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Transporter activity
- Specific Function:
- Aquaporins facilitate the transport of water and small neutral solutes across cell membranes.
- Gene Name:
- AQP12A
- Uniprot ID:
- Q8IXF9
- Molecular Weight:
- 31474.21 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Transporter activity
- Specific Function:
- Aquaporins facilitate the transport of water and small neutral solutes across cell membranes.
- Gene Name:
- AQP12B
- Uniprot ID:
- A6NM10
- Molecular Weight:
- 31475.13 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water transmembrane transporter activity
- Specific Function:
- Forms a water-specific channel that provides the plasma membranes of renal collecting duct with high permeability to water, thereby permitting water to move in the direction of an osmotic gradient.
- Gene Name:
- AQP2
- Uniprot ID:
- P41181
- Molecular Weight:
- 28837.17 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Water channel required to promote glycerol permeability and water transport across cell membranes. Acts as a glycerol transporter in skin and plays an important role in regulating SC (stratum corneum) and epidermal glycerol content. Involved in skin hydration, wound healing, and tumorigenesis. Provides kidney medullary collecting duct with high permeability to water, thereby permitting water to move in the direction of an osmotic gradient. Slightly permeable to urea and may function as a water and urea exit mechanism in antidiuresis in collecting duct cells. It may play an important role in gastrointestinal tract water transport and in glycerol metabolism (By similarity).
- Gene Name:
- AQP3
- Uniprot ID:
- Q92482
- Molecular Weight:
- 31543.605 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water transmembrane transporter activity
- Specific Function:
- Forms a water-specific channel. Osmoreceptor which regulates body water balance and mediates water flow within the central nervous system.
- Gene Name:
- AQP4
- Uniprot ID:
- P55087
- Molecular Weight:
- 34829.43 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Forms a water-specific channel. Implicated in the generation of saliva, tears, and pulmonary secretions. Required for TRPV4 activation by hypotonicity (PubMed:16571723). Together with TRPV4, controls regulatory volume decrease in salivary epithelial cells (PubMed:16571723).
- Gene Name:
- AQP5
- Uniprot ID:
- P55064
- Molecular Weight:
- 28291.89 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Forms a water-specific channel that participates in distinct physiological functions such as glomerular filtration, tubular endocytosis and acid-base metabolism.
- Gene Name:
- AQP6
- Uniprot ID:
- Q13520
- Molecular Weight:
- 29370.215 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Forms a channel for water and glycerol.
- Gene Name:
- AQP7
- Uniprot ID:
- O14520
- Molecular Weight:
- 37231.325 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water channel activity
- Specific Function:
- Forms a water-specific channel; mercury-sensitive. Not permeable to glycerol or urea.
- Gene Name:
- AQP8
- Uniprot ID:
- O94778
- Molecular Weight:
- 27381.01 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Water transmembrane transporter activity
- Specific Function:
- Forms a channel with a broad specificity. Mediates passage of a wide variety of non-charged solutes including carbamides, polyols, purines, and pyrimidines in a phloretin- and mercury-sensitive manner, whereas amino acids, cyclic sugars, Na(+), K(+), Cl(-), and deprotonated monocarboxylates are excluded. Also permeable to urea and glycerol.
- Gene Name:
- AQP9
- Uniprot ID:
- O43315
- Molecular Weight:
- 31430.77 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Not Available
- Specific Function:
- Keratin-binding protein required for epithelial cell polarization. Involved in apical junction complex (AJC) assembly via its interaction with PARD3. Required for ciliogenesis.
- Gene Name:
- FBF1
- Uniprot ID:
- Q8TES7
- Molecular Weight:
- 125445.19 Da
References
- Li Y, Yan XP, Chen C, Xia YL, Jiang Y: Human serum albumin-mercurial species interactions. J Proteome Res. 2007 Jun;6(6):2277-86. Epub 2007 May 10. [17489621 ]
- General Function:
- Water channel activity
- Specific Function:
- Water channel. Channel activity is down-regulated by CALM when cytoplasmic Ca(2+) levels are increased. May be responsible for regulating the osmolarity of the lens. Interactions between homotetramers from adjoining membranes may stabilize cell junctions in the eye lens core (By similarity).
- Gene Name:
- MIP
- Uniprot ID:
- P30301
- Molecular Weight:
- 28121.5 Da
References
- Yukutake Y, Tsuji S, Hirano Y, Adachi T, Takahashi T, Fujihara K, Agre P, Yasui M, Suematsu M: Mercury chloride decreases the water permeability of aquaporin-4-reconstituted proteoliposomes. Biol Cell. 2008 Jun;100(6):355-63. doi: 10.1042/BC20070132. [18167118 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Calcium-activated, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that is involved in positive and negative regulation of cell proliferation, apoptosis, differentiation, migration and adhesion, tumorigenesis, cardiac hypertrophy, angiogenesis, platelet function and inflammation, by directly phosphorylating targets such as RAF1, BCL2, CSPG4, TNNT2/CTNT, or activating signaling cascade involving MAPK1/3 (ERK1/2) and RAP1GAP. Involved in cell proliferation and cell growth arrest by positive and negative regulation of the cell cycle. Can promote cell growth by phosphorylating and activating RAF1, which mediates the activation of the MAPK/ERK signaling cascade, and/or by up-regulating CDKN1A, which facilitates active cyclin-dependent kinase (CDK) complex formation in glioma cells. In intestinal cells stimulated by the phorbol ester PMA, can trigger a cell cycle arrest program which is associated with the accumulation of the hyper-phosphorylated growth-suppressive form of RB1 and induction of the CDK inhibitors CDKN1A and CDKN1B. Exhibits anti-apoptotic function in glioma cells and protects them from apoptosis by suppressing the p53/TP53-mediated activation of IGFBP3, and in leukemia cells mediates anti-apoptotic action by phosphorylating BCL2. During macrophage differentiation induced by macrophage colony-stimulating factor (CSF1), is translocated to the nucleus and is associated with macrophage development. After wounding, translocates from focal contacts to lamellipodia and participates in the modulation of desmosomal adhesion. Plays a role in cell motility by phosphorylating CSPG4, which induces association of CSPG4 with extensive lamellipodia at the cell periphery and polarization of the cell accompanied by increases in cell motility. Is highly expressed in a number of cancer cells where it can act as a tumor promoter and is implicated in malignant phenotypes of several tumors such as gliomas and breast cancers. Negatively regulates myocardial contractility and positively regulates angiogenesis, platelet aggregation and thrombus formation in arteries. Mediates hypertrophic growth of neonatal cardiomyocytes, in part through a MAPK1/3 (ERK1/2)-dependent signaling pathway, and upon PMA treatment, is required to induce cardiomyocyte hypertrophy up to heart failure and death, by increasing protein synthesis, protein-DNA ratio and cell surface area. Regulates cardiomyocyte function by phosphorylating cardiac troponin T (TNNT2/CTNT), which induces significant reduction in actomyosin ATPase activity, myofilament calcium sensitivity and myocardial contractility. In angiogenesis, is required for full endothelial cell migration, adhesion to vitronectin (VTN), and vascular endothelial growth factor A (VEGFA)-dependent regulation of kinase activation and vascular tube formation. Involved in the stabilization of VEGFA mRNA at post-transcriptional level and mediates VEGFA-induced cell proliferation. In the regulation of calcium-induced platelet aggregation, mediates signals from the CD36/GP4 receptor for granule release, and activates the integrin heterodimer ITGA2B-ITGB3 through the RAP1GAP pathway for adhesion. During response to lipopolysaccharides (LPS), may regulate selective LPS-induced macrophage functions involved in host defense and inflammation. But in some inflammatory responses, may negatively regulate NF-kappa-B-induced genes, through IL1A-dependent induction of NF-kappa-B inhibitor alpha (NFKBIA/IKBA). Upon stimulation with 12-O-tetradecanoylphorbol-13-acetate (TPA), phosphorylates EIF4G1, which modulates EIF4G1 binding to MKNK1 and may be involved in the regulation of EIF4E phosphorylation. Phosphorylates KIT, leading to inhibition of KIT activity. Phosphorylates ATF2 which promotes cooperation between ATF2 and JUN, activating transcription.
- Gene Name:
- PRKCA
- Uniprot ID:
- P17252
- Molecular Weight:
- 76749.445 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Zinc ion binding
- Specific Function:
- Calcium-activated, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase involved in various cellular processes such as regulation of the B-cell receptor (BCR) signalosome, oxidative stress-induced apoptosis, androgen receptor-dependent transcription regulation, insulin signaling and endothelial cells proliferation. Plays a key role in B-cell activation by regulating BCR-induced NF-kappa-B activation. Mediates the activation of the canonical NF-kappa-B pathway (NFKB1) by direct phosphorylation of CARD11/CARMA1 at 'Ser-559', 'Ser-644' and 'Ser-652'. Phosphorylation induces CARD11/CARMA1 association with lipid rafts and recruitment of the BCL10-MALT1 complex as well as MAP3K7/TAK1, which then activates IKK complex, resulting in nuclear translocation and activation of NFKB1. Plays a direct role in the negative feedback regulation of the BCR signaling, by down-modulating BTK function via direct phosphorylation of BTK at 'Ser-180', which results in the alteration of BTK plasma membrane localization and in turn inhibition of BTK activity. Involved in apoptosis following oxidative damage: in case of oxidative conditions, specifically phosphorylates 'Ser-36' of isoform p66Shc of SHC1, leading to mitochondrial accumulation of p66Shc, where p66Shc acts as a reactive oxygen species producer. Acts as a coactivator of androgen receptor (ANDR)-dependent transcription, by being recruited to ANDR target genes and specifically mediating phosphorylation of 'Thr-6' of histone H3 (H3T6ph), a specific tag for epigenetic transcriptional activation that prevents demethylation of histone H3 'Lys-4' (H3K4me) by LSD1/KDM1A. In insulin signaling, may function downstream of IRS1 in muscle cells and mediate insulin-dependent DNA synthesis through the RAF1-MAPK/ERK signaling cascade. May participate in the regulation of glucose transport in adipocytes by negatively modulating the insulin-stimulated translocation of the glucose transporter SLC2A4/GLUT4. Under high glucose in pancreatic beta-cells, is probably involved in the inhibition of the insulin gene transcription, via regulation of MYC expression. In endothelial cells, activation of PRKCB induces increased phosphorylation of RB1, increased VEGFA-induced cell proliferation, and inhibits PI3K/AKT-dependent nitric oxide synthase (NOS3/eNOS) regulation by insulin, which causes endothelial dysfunction. Also involved in triglyceride homeostasis (By similarity). Phosphorylates ATF2 which promotes cooperation between ATF2 and JUN, activating transcription.
- Gene Name:
- PRKCB
- Uniprot ID:
- P05771
- Molecular Weight:
- 76868.45 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Protein serine/threonine kinase activity
- Specific Function:
- Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays contrasting roles in cell death and cell survival by functioning as a pro-apoptotic protein during DNA damage-induced apoptosis, but acting as an anti-apoptotic protein during cytokine receptor-initiated cell death, is involved in tumor suppression as well as survival of several cancers, is required for oxygen radical production by NADPH oxidase and acts as positive or negative regulator in platelet functional responses. Negatively regulates B cell proliferation and also has an important function in self-antigen induced B cell tolerance induction. Upon DNA damage, activates the promoter of the death-promoting transcription factor BCLAF1/Btf to trigger BCLAF1-mediated p53/TP53 gene transcription and apoptosis. In response to oxidative stress, interact with and activate CHUK/IKKA in the nucleus, causing the phosphorylation of p53/TP53. In the case of ER stress or DNA damage-induced apoptosis, can form a complex with the tyrosine-protein kinase ABL1 which trigger apoptosis independently of p53/TP53. In cytosol can trigger apoptosis by activating MAPK11 or MAPK14, inhibiting AKT1 and decreasing the level of X-linked inhibitor of apoptosis protein (XIAP), whereas in nucleus induces apoptosis via the activation of MAPK8 or MAPK9. Upon ionizing radiation treatment, is required for the activation of the apoptosis regulators BAX and BAK, which trigger the mitochondrial cell death pathway. Can phosphorylate MCL1 and target it for degradation which is sufficient to trigger for BAX activation and apoptosis. Is required for the control of cell cycle progression both at G1/S and G2/M phases. Mediates phorbol 12-myristate 13-acetate (PMA)-induced inhibition of cell cycle progression at G1/S phase by up-regulating the CDK inhibitor CDKN1A/p21 and inhibiting the cyclin CCNA2 promoter activity. In response to UV irradiation can phosphorylate CDK1, which is important for the G2/M DNA damage checkpoint activation. Can protect glioma cells from the apoptosis induced by TNFSF10/TRAIL, probably by inducing increased phosphorylation and subsequent activation of AKT1. Is highly expressed in a number of cancer cells and promotes cell survival and resistance against chemotherapeutic drugs by inducing cyclin D1 (CCND1) and hyperphosphorylation of RB1, and via several pro-survival pathways, including NF-kappa-B, AKT1 and MAPK1/3 (ERK1/2). Can also act as tumor suppressor upon mitogenic stimulation with PMA or TPA. In N-formyl-methionyl-leucyl-phenylalanine (fMLP)-treated cells, is required for NCF1 (p47-phox) phosphorylation and activation of NADPH oxidase activity, and regulates TNF-elicited superoxide anion production in neutrophils, by direct phosphorylation and activation of NCF1 or indirectly through MAPK1/3 (ERK1/2) signaling pathways. May also play a role in the regulation of NADPH oxidase activity in eosinophil after stimulation with IL5, leukotriene B4 or PMA. In collagen-induced platelet aggregation, acts a negative regulator of filopodia formation and actin polymerization by interacting with and negatively regulating VASP phosphorylation. Downstream of PAR1, PAR4 and CD36/GP4 receptors, regulates differentially platelet dense granule secretion; acts as a positive regulator in PAR-mediated granule secretion, whereas it negatively regulates CD36/GP4-mediated granule release. Phosphorylates MUC1 in the C-terminal and regulates the interaction between MUC1 and beta-catenin. The catalytic subunit phosphorylates 14-3-3 proteins (YWHAB, YWHAZ and YWHAH) in a sphingosine-dependent fashion (By similarity).
- Gene Name:
- PRKCD
- Uniprot ID:
- Q05655
- Molecular Weight:
- 77504.445 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Signal transducer activity
- Specific Function:
- Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays essential roles in the regulation of multiple cellular processes linked to cytoskeletal proteins, such as cell adhesion, motility, migration and cell cycle, functions in neuron growth and ion channel regulation, and is involved in immune response, cancer cell invasion and regulation of apoptosis. Mediates cell adhesion to the extracellular matrix via integrin-dependent signaling, by mediating angiotensin-2-induced activation of integrin beta-1 (ITGB1) in cardiac fibroblasts. Phosphorylates MARCKS, which phosphorylates and activates PTK2/FAK, leading to the spread of cardiomyocytes. Involved in the control of the directional transport of ITGB1 in mesenchymal cells by phosphorylating vimentin (VIM), an intermediate filament (IF) protein. In epithelial cells, associates with and phosphorylates keratin-8 (KRT8), which induces targeting of desmoplakin at desmosomes and regulates cell-cell contact. Phosphorylates IQGAP1, which binds to CDC42, mediating epithelial cell-cell detachment prior to migration. In HeLa cells, contributes to hepatocyte growth factor (HGF)-induced cell migration, and in human corneal epithelial cells, plays a critical role in wound healing after activation by HGF. During cytokinesis, forms a complex with YWHAB, which is crucial for daughter cell separation, and facilitates abscission by a mechanism which may implicate the regulation of RHOA. In cardiac myocytes, regulates myofilament function and excitation coupling at the Z-lines, where it is indirectly associated with F-actin via interaction with COPB1. During endothelin-induced cardiomyocyte hypertrophy, mediates activation of PTK2/FAK, which is critical for cardiomyocyte survival and regulation of sarcomere length. Plays a role in the pathogenesis of dilated cardiomyopathy via persistent phosphorylation of troponin I (TNNI3). Involved in nerve growth factor (NFG)-induced neurite outgrowth and neuron morphological change independently of its kinase activity, by inhibition of RHOA pathway, activation of CDC42 and cytoskeletal rearrangement. May be involved in presynaptic facilitation by mediating phorbol ester-induced synaptic potentiation. Phosphorylates gamma-aminobutyric acid receptor subunit gamma-2 (GABRG2), which reduces the response of GABA receptors to ethanol and benzodiazepines and may mediate acute tolerance to the intoxicating effects of ethanol. Upon PMA treatment, phosphorylates the capsaicin- and heat-activated cation channel TRPV1, which is required for bradykinin-induced sensitization of the heat response in nociceptive neurons. Is able to form a complex with PDLIM5 and N-type calcium channel, and may enhance channel activities and potentiates fast synaptic transmission by phosphorylating the pore-forming alpha subunit CACNA1B (CaV2.2). In prostate cancer cells, interacts with and phosphorylates STAT3, which increases DNA-binding and transcriptional activity of STAT3 and seems to be essential for prostate cancer cell invasion. Downstream of TLR4, plays an important role in the lipopolysaccharide (LPS)-induced immune response by phosphorylating and activating TICAM2/TRAM, which in turn activates the transcription factor IRF3 and subsequent cytokines production. In differentiating erythroid progenitors, is regulated by EPO and controls the protection against the TNFSF10/TRAIL-mediated apoptosis, via BCL2. May be involved in the regulation of the insulin-induced phosphorylation and activation of AKT1.
- Gene Name:
- PRKCE
- Uniprot ID:
- Q02156
- Molecular Weight:
- 83673.2 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Protein kinase c activity
- Specific Function:
- Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that is involved in the regulation of cell differentiation in keratinocytes and pre-B cell receptor, mediates regulation of epithelial tight junction integrity and foam cell formation, and is required for glioblastoma proliferation and apoptosis prevention in MCF-7 cells. In keratinocytes, binds and activates the tyrosine kinase FYN, which in turn blocks epidermal growth factor receptor (EGFR) signaling and leads to keratinocyte growth arrest and differentiation. Associates with the cyclin CCNE1-CDK2-CDKN1B complex and inhibits CDK2 kinase activity, leading to RB1 dephosphorylation and thereby G1 arrest in keratinocytes. In association with RALA activates actin depolymerization, which is necessary for keratinocyte differentiation. In the pre-B cell receptor signaling, functions downstream of BLNK by up-regulating IRF4, which in turn activates L chain gene rearrangement. Regulates epithelial tight junctions (TJs) by phosphorylating occludin (OCLN) on threonine residues, which is necessary for the assembly and maintenance of TJs. In association with PLD2 and via TLR4 signaling, is involved in lipopolysaccharide (LPS)-induced RGS2 down-regulation and foam cell formation. Upon PMA stimulation, mediates glioblastoma cell proliferation by activating the mTOR pathway, the PI3K/AKT pathway and the ERK1-dependent phosphorylation of ELK1. Involved in the protection of glioblastoma cells from irradiation-induced apoptosis by preventing caspase-9 activation. In camptothecin-treated MCF-7 cells, regulates NF-kappa-B upstream signaling by activating IKBKB, and confers protection against DNA damage-induced apoptosis. Promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Phosphorylates ATF2 which promotes its nuclear retention and transcriptional activity and negatively regulates its mitochondrial localization.
- Gene Name:
- PRKCH
- Uniprot ID:
- P24723
- Molecular Weight:
- 77827.96 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Zinc ion binding
- Specific Function:
- Calcium-activated, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays diverse roles in neuronal cells and eye tissues, such as regulation of the neuronal receptors GRIA4/GLUR4 and GRIN1/NMDAR1, modulation of receptors and neuronal functions related to sensitivity to opiates, pain and alcohol, mediation of synaptic function and cell survival after ischemia, and inhibition of gap junction activity after oxidative stress. Binds and phosphorylates GRIA4/GLUR4 glutamate receptor and regulates its function by increasing plasma membrane-associated GRIA4 expression. In primary cerebellar neurons treated with the agonist 3,5-dihyidroxyphenylglycine, functions downstream of the metabotropic glutamate receptor GRM5/MGLUR5 and phosphorylates GRIN1/NMDAR1 receptor which plays a key role in synaptic plasticity, synaptogenesis, excitotoxicity, memory acquisition and learning. May be involved in the regulation of hippocampal long-term potentiation (LTP), but may be not necessary for the process of synaptic plasticity. May be involved in desensitization of mu-type opioid receptor-mediated G-protein activation in the spinal cord, and may be critical for the development and/or maintenance of morphine-induced reinforcing effects in the limbic forebrain. May modulate the functionality of mu-type-opioid receptors by participating in a signaling pathway which leads to the phosphorylation and degradation of opioid receptors. May also contributes to chronic morphine-induced changes in nociceptive processing. Plays a role in neuropathic pain mechanisms and contributes to the maintenance of the allodynia pain produced by peripheral inflammation. Plays an important role in initial sensitivity and tolerance to ethanol, by mediating the behavioral effects of ethanol as well as the effects of this drug on the GABA(A) receptors. During and after cerebral ischemia modulate neurotransmission and cell survival in synaptic membranes, and is involved in insulin-induced inhibition of necrosis, an important mechanism for minimizing ischemic injury. Required for the elimination of multiple climbing fibers during innervation of Purkinje cells in developing cerebellum. Is activated in lens epithelial cells upon hydrogen peroxide treatment, and phosphorylates connexin-43 (GJA1/CX43), resulting in disassembly of GJA1 gap junction plaques and inhibition of gap junction activity which could provide a protective effect against oxidative stress (By similarity). Phosphorylates p53/TP53 and promotes p53/TP53-dependent apoptosis in response to DNA damage. Involved in the phase resetting of the cerebral cortex circadian clock during temporally restricted feeding. Stabilizes the core clock component ARNTL/BMAL1 by interfering with its ubiquitination, thus suppressing its degradation, resulting in phase resetting of the cerebral cortex clock (By similarity).
- Gene Name:
- PRKCG
- Uniprot ID:
- P05129
- Molecular Weight:
- 78447.23 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Protein serine/threonine kinase activity
- Specific Function:
- Calcium- and diacylglycerol-independent serine/ threonine-protein kinase that plays a general protective role against apoptotic stimuli, is involved in NF-kappa-B activation, cell survival, differentiation and polarity, and contributes to the regulation of microtubule dynamics in the early secretory pathway. Is necessary for BCR-ABL oncogene-mediated resistance to apoptotic drug in leukemia cells, protecting leukemia cells against drug-induced apoptosis. In cultured neurons, prevents amyloid beta protein-induced apoptosis by interrupting cell death process at a very early step. In glioblastoma cells, may function downstream of phosphatidylinositol 3-kinase (PI(3)K) and PDPK1 in the promotion of cell survival by phosphorylating and inhibiting the pro-apoptotic factor BAD. Can form a protein complex in non-small cell lung cancer (NSCLC) cells with PARD6A and ECT2 and regulate ECT2 oncogenic activity by phosphorylation, which in turn promotes transformed growth and invasion. In response to nerve growth factor (NGF), acts downstream of SRC to phosphorylate and activate IRAK1, allowing the subsequent activation of NF-kappa-B and neuronal cell survival. Functions in the organization of the apical domain in epithelial cells by phosphorylating EZR. This step is crucial for activation and normal distribution of EZR at the early stages of intestinal epithelial cell differentiation. Forms a protein complex with LLGL1 and PARD6B independently of PARD3 to regulate epithelial cell polarity. Plays a role in microtubule dynamics in the early secretory pathway through interaction with RAB2A and GAPDH and recruitment to vesicular tubular clusters (VTCs). In human coronary artery endothelial cells (HCAEC), is activated by saturated fatty acids and mediates lipid-induced apoptosis.
- Gene Name:
- PRKCI
- Uniprot ID:
- P41743
- Molecular Weight:
- 68261.855 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Ubiquitin-protein transferase activity
- Specific Function:
- Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that mediates non-redundant functions in T-cell receptor (TCR) signaling, including T-cells activation, proliferation, differentiation and survival, by mediating activation of multiple transcription factors such as NF-kappa-B, JUN, NFATC1 and NFATC2. In TCR-CD3/CD28-co-stimulated T-cells, is required for the activation of NF-kappa-B and JUN, which in turn are essential for IL2 production, and participates to the calcium-dependent NFATC1 and NFATC2 transactivation. Mediates the activation of the canonical NF-kappa-B pathway (NFKB1) by direct phosphorylation of CARD11 on several serine residues, inducing CARD11 association with lipid rafts and recruitment of the BCL10-MALT1 complex, which then activates IKK complex, resulting in nuclear translocation and activation of NFKB1. May also play an indirect role in activation of the non-canonical NF-kappa-B (NFKB2) pathway. In the signaling pathway leading to JUN activation, acts by phosphorylating the mediator STK39/SPAK and may not act through MAP kinases signaling. Plays a critical role in TCR/CD28-induced NFATC1 and NFATC2 transactivation by participating in the regulation of reduced inositol 1,4,5-trisphosphate generation and intracellular calcium mobilization. After costimulation of T-cells through CD28 can phosphorylate CBLB and is required for the ubiquitination and subsequent degradation of CBLB, which is a prerequisite for the activation of TCR. During T-cells differentiation, plays an important role in the development of T-helper 2 (Th2) cells following immune and inflammatory responses, and, in the development of inflammatory autoimmune diseases, is necessary for the activation of IL17-producing Th17 cells. May play a minor role in Th1 response. Upon TCR stimulation, mediates T-cell protective survival signal by phosphorylating BAD, thus protecting T-cells from BAD-induced apoptosis, and by up-regulating BCL-X(L)/BCL2L1 levels through NF-kappa-B and JUN pathways. In platelets, regulates signal transduction downstream of the ITGA2B, CD36/GP4, F2R/PAR1 and F2RL3/PAR4 receptors, playing a positive role in 'outside-in' signaling and granule secretion signal transduction. May relay signals from the activated ITGA2B receptor by regulating the uncoupling of WASP and WIPF1, thereby permitting the regulation of actin filament nucleation and branching activity of the Arp2/3 complex. May mediate inhibitory effects of free fatty acids on insulin signaling by phosphorylating IRS1, which in turn blocks IRS1 tyrosine phosphorylation and downstream activation of the PI3K/AKT pathway. Phosphorylates MSN (moesin) in the presence of phosphatidylglycerol or phosphatidylinositol. Phosphorylates PDPK1 at 'Ser-504' and 'Ser-532' and negatively regulates its ability to phosphorylate PKB/AKT1.
- Gene Name:
- PRKCQ
- Uniprot ID:
- Q04759
- Molecular Weight:
- 81864.145 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Protein serine/threonine kinase activity
- Specific Function:
- Calcium- and diacylglycerol-independent serine/threonine-protein kinase that functions in phosphatidylinositol 3-kinase (PI3K) pathway and mitogen-activated protein (MAP) kinase cascade, and is involved in NF-kappa-B activation, mitogenic signaling, cell proliferation, cell polarity, inflammatory response and maintenance of long-term potentiation (LTP). Upon lipopolysaccharide (LPS) treatment in macrophages, or following mitogenic stimuli, functions downstream of PI3K to activate MAP2K1/MEK1-MAPK1/ERK2 signaling cascade independently of RAF1 activation. Required for insulin-dependent activation of AKT3, but may function as an adapter rather than a direct activator. Upon insulin treatment may act as a downstream effector of PI3K and contribute to the activation of translocation of the glucose transporter SLC2A4/GLUT4 and subsequent glucose transport in adipocytes. In EGF-induced cells, binds and activates MAP2K5/MEK5-MAPK7/ERK5 independently of its kinase activity and can activate JUN promoter through MEF2C. Through binding with SQSTM1/p62, functions in interleukin-1 signaling and activation of NF-kappa-B with the specific adapters RIPK1 and TRAF6. Participates in TNF-dependent transactivation of NF-kappa-B by phosphorylating and activating IKBKB kinase, which in turn leads to the degradation of NF-kappa-B inhibitors. In migrating astrocytes, forms a cytoplasmic complex with PARD6A and is recruited by CDC42 to function in the establishment of cell polarity along with the microtubule motor and dynein. In association with FEZ1, stimulates neuronal differentiation in PC12 cells. In the inflammatory response, is required for the T-helper 2 (Th2) differentiation process, including interleukin production, efficient activation of JAK1 and the subsequent phosphorylation and nuclear translocation of STAT6. May be involved in development of allergic airway inflammation (asthma), a process dependent on Th2 immune response. In the NF-kappa-B-mediated inflammatory response, can relieve SETD6-dependent repression of NF-kappa-B target genes by phosphorylating the RELA subunit at 'Ser-311'. Necessary and sufficient for LTP maintenance in hippocampal CA1 pyramidal cells. In vein endothelial cells treated with the oxidant peroxynitrite, phosphorylates STK11 leading to nuclear export of STK11, subsequent inhibition of PI3K/Akt signaling, and increased apoptosis. Phosphorylates VAMP2 in vitro (PubMed:17313651).
- Gene Name:
- PRKCZ
- Uniprot ID:
- Q05513
- Molecular Weight:
- 67659.335 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- Not Available
- Uniprot ID:
- A6NKZ8
- Molecular Weight:
- Not Available
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- Not Available
- Uniprot ID:
- Q99867
- Molecular Weight:
- Not Available
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Not Available
- Gene Name:
- TUBA4B
- Uniprot ID:
- Q9H853
- Molecular Weight:
- 27551.01 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBAL3
- Uniprot ID:
- A6NHL2
- Molecular Weight:
- 49908.305 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Specific Function:
- Gtp binding
- Gene Name:
- TUBA1A
- Uniprot ID:
- Q71U36
- Molecular Weight:
- 50135.25 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBA1B
- Uniprot ID:
- P68363
- Molecular Weight:
- 50151.24 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural molecule activity
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBA1C
- Uniprot ID:
- Q9BQE3
- Molecular Weight:
- 49894.93 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBA3C
- Uniprot ID:
- Q13748
- Molecular Weight:
- 49959.145 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBA3E
- Uniprot ID:
- Q6PEY2
- Molecular Weight:
- 49858.135 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBA4A
- Uniprot ID:
- P68366
- Molecular Weight:
- 49923.995 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Specific Function:
- Gtp binding
- Gene Name:
- TUBA8
- Uniprot ID:
- Q9NY65
- Molecular Weight:
- 50093.12 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBB
- Uniprot ID:
- P07437
- Molecular Weight:
- 49670.515 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBB1
- Uniprot ID:
- Q9H4B7
- Molecular Weight:
- 50326.56 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBB2A
- Uniprot ID:
- Q13885
- Molecular Weight:
- 49906.67 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity). TUBB2B is implicated in neuronal migration.
- Gene Name:
- TUBB2B
- Uniprot ID:
- Q9BVA1
- Molecular Weight:
- 49952.76 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain. TUBB3 plays a critical role in proper axon guidance and mantainance.
- Gene Name:
- TUBB3
- Uniprot ID:
- Q13509
- Molecular Weight:
- 50432.355 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBB4A
- Uniprot ID:
- P04350
- Molecular Weight:
- 49585.475 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Unfolded protein binding
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Gene Name:
- TUBB4B
- Uniprot ID:
- P68371
- Molecular Weight:
- 49830.72 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBB6
- Uniprot ID:
- Q9BUF5
- Molecular Weight:
- 49856.785 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- TUBB8
- Uniprot ID:
- Q3ZCM7
- Molecular Weight:
- 49775.655 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain (By similarity).
- Gene Name:
- Not Available
- Uniprot ID:
- A6NNZ2
- Molecular Weight:
- 49572.265 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- In the elongating spermatid it is associated with the manchette, a specialized microtubule system present during reshaping of the sperm head.
- Gene Name:
- TUBD1
- Uniprot ID:
- Q9UJT1
- Molecular Weight:
- 51033.86 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Not Available
- Gene Name:
- TUBE1
- Uniprot ID:
- Q9UJT0
- Molecular Weight:
- 52931.4 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Tubulin is the major constituent of microtubules. The gamma chain is found at microtubule organizing centers (MTOC) such as the spindle poles or the centrosome. Pericentriolar matrix component that regulates alpha/beta chain minus-end nucleation, centrosome duplication and spindle formation.
- Gene Name:
- TUBG1
- Uniprot ID:
- P23258
- Molecular Weight:
- 51169.48 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Structural molecule activity
- Specific Function:
- Tubulin is the major constituent of microtubules. The gamma chain is found at microtubule organizing centers (MTOC) such as the spindle poles or the centrosome. Pericentriolar matrix component that regulates alpha/beta chain minus-end nucleation, centrosome duplication and spindle formation (By similarity).
- Gene Name:
- TUBG2
- Uniprot ID:
- Q9NRH3
- Molecular Weight:
- 51091.32 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for mercury. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Sh2 domain binding
- Specific Function:
- Non-receptor tyrosine-protein kinase that plays an essential role in the selection and maturation of developing T-cells in the thymus and in the function of mature T-cells. Plays a key role in T-cell antigen receptor (TCR)-linked signal transduction pathways. Constitutively associated with the cytoplasmic portions of the CD4 and CD8 surface receptors. Association of the TCR with a peptide antigen-bound MHC complex facilitates the interaction of CD4 and CD8 with MHC class II and class I molecules, respectively, thereby recruiting the associated LCK protein to the vicinity of the TCR/CD3 complex. LCK then phosphorylates tyrosines residues within the immunoreceptor tyrosine-based activation motifs (ITAM) of the cytoplasmic tails of the TCR-gamma chains and CD3 subunits, initiating the TCR/CD3 signaling pathway. Once stimulated, the TCR recruits the tyrosine kinase ZAP70, that becomes phosphorylated and activated by LCK. Following this, a large number of signaling molecules are recruited, ultimately leading to lymphokine production. LCK also contributes to signaling by other receptor molecules. Associates directly with the cytoplasmic tail of CD2, which leads to hyperphosphorylation and activation of LCK. Also plays a role in the IL2 receptor-linked signaling pathway that controls the T-cell proliferative response. Binding of IL2 to its receptor results in increased activity of LCK. Is expressed at all stages of thymocyte development and is required for the regulation of maturation events that are governed by both pre-TCR and mature alpha beta TCR. Phosphorylates other substrates including RUNX3, PTK2B/PYK2, the microtubule-associated protein MAPT, RHOH or TYROBP.
- Gene Name:
- LCK
- Uniprot ID:
- P06239
- Molecular Weight:
- 58000.15 Da
References
- Ziemba SE, Menard SL, McCabe MJ Jr, Rosenspire AJ: T-cell receptor signaling is mediated by transient Lck activity, which is inhibited by inorganic mercury. FASEB J. 2009 Jun;23(6):1663-71. doi: 10.1096/fj.08-117283. Epub 2009 Jan 23. [19168706 ]
- General Function:
- Voltage-gated calcium channel activity
- Specific Function:
- Receptor for endogenous opioids such as beta-endorphin and endomorphin. Receptor for natural and synthetic opioids including morphine, heroin, DAMGO, fentanyl, etorphine, buprenorphin and methadone. Agonist binding to the receptor induces coupling to an inactive GDP-bound heterotrimeric G-protein complex and subsequent exchange of GDP for GTP in the G-protein alpha subunit leading to dissociation of the G-protein complex with the free GTP-bound G-protein alpha and the G-protein beta-gamma dimer activating downstream cellular effectors. The agonist- and cell type-specific activity is predominantly coupled to pertussis toxin-sensitive G(i) and G(o) G alpha proteins, GNAI1, GNAI2, GNAI3 and GNAO1 isoforms Alpha-1 and Alpha-2, and to a lesser extend to pertussis toxin-insensitive G alpha proteins GNAZ and GNA15. They mediate an array of downstream cellular responses, including inhibition of adenylate cyclase activity and both N-type and L-type calcium channels, activation of inward rectifying potassium channels, mitogen-activated protein kinase (MAPK), phospholipase C (PLC), phosphoinositide/protein kinase (PKC), phosphoinositide 3-kinase (PI3K) and regulation of NF-kappa-B. Also couples to adenylate cyclase stimulatory G alpha proteins. The selective temporal coupling to G-proteins and subsequent signaling can be regulated by RGSZ proteins, such as RGS9, RGS17 and RGS4. Phosphorylation by members of the GPRK subfamily of Ser/Thr protein kinases and association with beta-arrestins is involved in short-term receptor desensitization. Beta-arrestins associate with the GPRK-phosphorylated receptor and uncouple it from the G-protein thus terminating signal transduction. The phosphorylated receptor is internalized through endocytosis via clathrin-coated pits which involves beta-arrestins. The activation of the ERK pathway occurs either in a G-protein-dependent or a beta-arrestin-dependent manner and is regulated by agonist-specific receptor phosphorylation. Acts as a class A G-protein coupled receptor (GPCR) which dissociates from beta-arrestin at or near the plasma membrane and undergoes rapid recycling. Receptor down-regulation pathways are varying with the agonist and occur dependent or independent of G-protein coupling. Endogenous ligands induce rapid desensitization, endocytosis and recycling whereas morphine induces only low desensitization and endocytosis. Heterooligomerization with other GPCRs can modulate agonist binding, signaling and trafficking properties. Involved in neurogenesis. Isoform 12 couples to GNAS and is proposed to be involved in excitatory effects. Isoform 16 and isoform 17 do not bind agonists but may act through oligomerization with binding-competent OPRM1 isoforms and reduce their ligand binding activity.
- Gene Name:
- OPRM1
- Uniprot ID:
- P35372
- Molecular Weight:
- 44778.855 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.18 uM | NVS_GPCR_hOpiate_mu | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- G-protein coupled amine receptor activity
- Specific Function:
- Dopamine receptor whose activity is mediated by G proteins which activate adenylyl cyclase.
- Gene Name:
- DRD1
- Uniprot ID:
- P21728
- Molecular Weight:
- 49292.765 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.19 uM | NVS_GPCR_hDRD1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein homodimerization activity
- Specific Function:
- Alpha-2 adrenergic receptors mediate the catecholamine-induced inhibition of adenylate cyclase through the action of G proteins.
- Gene Name:
- ADRA2C
- Uniprot ID:
- P18825
- Molecular Weight:
- 49521.585 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.23 uM | NVS_GPCR_hAdra2C | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Opioid receptor activity
- Specific Function:
- G-protein coupled receptor that functions as receptor for endogenous enkephalins and for a subset of other opioids. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling leads to the inhibition of adenylate cyclase activity. Inhibits neurotransmitter release by reducing calcium ion currents and increasing potassium ion conductance. Plays a role in the perception of pain and in opiate-mediated analgesia. Plays a role in developing analgesic tolerance to morphine.
- Gene Name:
- OPRD1
- Uniprot ID:
- P41143
- Molecular Weight:
- 40368.235 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.26 uM | NVS_GPCR_hOpiate_D1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor signaling protein activity
- Specific Function:
- Beta-adrenergic receptors mediate the catecholamine-induced activation of adenylate cyclase through the action of G proteins. This receptor binds epinephrine and norepinephrine with approximately equal affinity. Mediates Ras activation through G(s)-alpha- and cAMP-mediated signaling.
- Gene Name:
- ADRB1
- Uniprot ID:
- P08588
- Molecular Weight:
- 51322.1 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.30 uM | NVS_GPCR_hAdrb1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Nociceptin receptor activity
- Specific Function:
- G-protein coupled opioid receptor that functions as receptor for the endogenous neuropeptide nociceptin. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors. Signaling via G proteins mediates inhibition of adenylate cyclase activity and calcium channel activity. Arrestins modulate signaling via G proteins and mediate the activation of alternative signaling pathways that lead to the activation of MAP kinases. Plays a role in modulating nociception and the perception of pain. Plays a role in the regulation of locomotor activity by the neuropeptide nociceptin.
- Gene Name:
- OPRL1
- Uniprot ID:
- P41146
- Molecular Weight:
- 40692.775 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.31 uM | NVS_GPCR_hORL1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Potassium channel regulator activity
- Specific Function:
- Dopamine receptor whose activity is mediated by G proteins which inhibit adenylyl cyclase.
- Gene Name:
- DRD2
- Uniprot ID:
- P14416
- Molecular Weight:
- 50618.91 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.35 uM | NVS_GPCR_hDRD2s | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Phosphatidylinositol phospholipase c activity
- Specific Function:
- The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. Primary transducing effect is Pi turnover.
- Gene Name:
- CHRM5
- Uniprot ID:
- P08912
- Molecular Weight:
- 60073.205 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.35 uM | NVS_GPCR_hM5 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Norepinephrine:sodium symporter activity
- Specific Function:
- Amine transporter. Terminates the action of noradrenaline by its high affinity sodium-dependent reuptake into presynaptic terminals.
- Gene Name:
- SLC6A2
- Uniprot ID:
- P23975
- Molecular Weight:
- 69331.42 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.36 uM | NVS_TR_hNET | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor activity
- Specific Function:
- The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. Primary transducing effect is Pi turnover.
- Gene Name:
- CHRM3
- Uniprot ID:
- P20309
- Molecular Weight:
- 66127.445 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.49 uM | NVS_GPCR_hM3 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Responsible for the metabolism of a number of therapeutic agents such as the anticonvulsant drug S-mephenytoin, omeprazole, proguanil, certain barbiturates, diazepam, propranolol, citalopram and imipramine.
- Gene Name:
- CYP2C19
- Uniprot ID:
- P33261
- Molecular Weight:
- 55930.545 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.80 uM | NVS_ADME_hCYP2C19 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins that stimulate adenylate cyclase.
- Gene Name:
- HTR7
- Uniprot ID:
- P34969
- Molecular Weight:
- 53554.43 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.83 uM | NVS_GPCR_h5HT7 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity).
- Gene Name:
- PPARG
- Uniprot ID:
- P37231
- Molecular Weight:
- 57619.58 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.86 uM | Tox21_PPARg_BLA_Agonist_ratio | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein homodimerization activity
- Specific Function:
- Beta-adrenergic receptors mediate the catecholamine-induced activation of adenylate cyclase through the action of G proteins. The beta-2-adrenergic receptor binds epinephrine with an approximately 30-fold greater affinity than it does norepinephrine.
- Gene Name:
- ADRB2
- Uniprot ID:
- P07550
- Molecular Weight:
- 46458.32 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.92 uM | NVS_GPCR_hAdrb2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Histamine receptor activity
- Specific Function:
- In peripheral tissues, the H1 subclass of histamine receptors mediates the contraction of smooth muscles, increase in capillary permeability due to contraction of terminal venules, and catecholamine release from adrenal medulla, as well as mediating neurotransmission in the central nervous system.
- Gene Name:
- HRH1
- Uniprot ID:
- P35367
- Molecular Weight:
- 55783.61 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.94 uM | NVS_GPCR_hH1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.99 uM | NVS_ADME_hCYP3A4 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Most active in catalyzing 2-hydroxylation. Caffeine is metabolized primarily by cytochrome CYP1A2 in the liver through an initial N3-demethylation. Also acts in the metabolism of aflatoxin B1 and acetaminophen. Participates in the bioactivation of carcinogenic aromatic and heterocyclic amines. Catalizes the N-hydroxylation of heterocyclic amines and the O-deethylation of phenacetin.
- Gene Name:
- CYP1A2
- Uniprot ID:
- P05177
- Molecular Weight:
- 58293.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.04 uM | NVS_ADME_hCYP1A2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Phosphatidylinositol phospholipase c activity
- Specific Function:
- The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. Primary transducing effect is Pi turnover.
- Gene Name:
- CHRM1
- Uniprot ID:
- P11229
- Molecular Weight:
- 51420.375 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.10 uM | NVS_GPCR_hM1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- G-protein coupled acetylcholine receptor activity
- Specific Function:
- The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. Primary transducing effect is adenylate cyclase inhibition. Signaling promotes phospholipase C activity, leading to the release of inositol trisphosphate (IP3); this then triggers calcium ion release into the cytosol.
- Gene Name:
- CHRM2
- Uniprot ID:
- P08172
- Molecular Weight:
- 51714.605 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.10 uM | NVS_GPCR_hM2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Steroid hormone receptors are ligand-activated transcription factors that regulate eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Transcription factor activity is modulated by bound coactivator and corepressor proteins. Transcription activation is down-regulated by NR0B2. Activated, but not phosphorylated, by HIPK3 and ZIPK/DAPK3.
- Gene Name:
- AR
- Uniprot ID:
- P10275
- Molecular Weight:
- 98987.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.45 uM | Tox21_AR_BLA_Agonist_ratio | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins.
- Gene Name:
- HTR5A
- Uniprot ID:
- P47898
- Molecular Weight:
- 40254.69 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.54 uM | NVS_GPCR_h5HT5A | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Sh3 domain binding
- Specific Function:
- Dopamine receptor responsible for neuronal signaling in the mesolimbic system of the brain, an area of the brain that regulates emotion and complex behavior. Its activity is mediated by G proteins which inhibit adenylyl cyclase. Modulates the circadian rhythm of contrast sensitivity by regulating the rhythmic expression of NPAS2 in the retinal ganglion cells (By similarity).
- Gene Name:
- DRD4
- Uniprot ID:
- P21917
- Molecular Weight:
- 48359.86 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.55 uM | NVS_GPCR_hDRD4.4 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins that stimulate adenylate cyclase. It has a high affinity for tricyclic psychotropic drugs (By similarity). Controls pyramidal neurons migration during corticogenesis, through the regulation of CDK5 activity (By similarity). Is an activator of TOR signaling (PubMed:23027611).
- Gene Name:
- HTR6
- Uniprot ID:
- P50406
- Molecular Weight:
- 46953.625 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.61 uM | NVS_GPCR_h5HT6 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Thioesterase binding
- Specific Function:
- Alpha-2 adrenergic receptors mediate the catecholamine-induced inhibition of adenylate cyclase through the action of G proteins. The rank order of potency for agonists of this receptor is oxymetazoline > clonidine > epinephrine > norepinephrine > phenylephrine > dopamine > p-synephrine > p-tyramine > serotonin = p-octopamine. For antagonists, the rank order is yohimbine > phentolamine = mianserine > chlorpromazine = spiperone = prazosin > propanolol > alprenolol = pindolol.
- Gene Name:
- ADRA2A
- Uniprot ID:
- P08913
- Molecular Weight:
- 48956.275 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 2.25 uM | NVS_GPCR_hAdra2A | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Responsible for the metabolism of many drugs and environmental chemicals that it oxidizes. It is involved in the metabolism of drugs such as antiarrhythmics, adrenoceptor antagonists, and tricyclic antidepressants.
- Gene Name:
- CYP2D6
- Uniprot ID:
- P10635
- Molecular Weight:
- 55768.94 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 2.45 uM | NVS_ADME_hCYP2D6 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Phosphatidylinositol phospholipase c activity
- Specific Function:
- Receptor for endothelin-1. Mediates its action by association with G proteins that activate a phosphatidylinositol-calcium second messenger system. The rank order of binding affinities for ET-A is: ET1 > ET2 >> ET3.
- Gene Name:
- EDNRA
- Uniprot ID:
- P25101
- Molecular Weight:
- 48721.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 2.96 uM | NVS_GPCR_hETA | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Tachykinin receptor activity
- Specific Function:
- This is a receptor for the tachykinin neuropeptide substance K (neurokinin A). It is associated with G proteins that activate a phosphatidylinositol-calcium second messenger system. The rank order of affinity of this receptor to tachykinins is: substance K > neuromedin-K > substance P.
- Gene Name:
- TACR2
- Uniprot ID:
- P21452
- Molecular Weight:
- 44441.705 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 3.41 uM | NVS_GPCR_hNK2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for glucocorticoids (GC). Has a dual mode of action: as a transcription factor that binds to glucocorticoid response elements (GRE), both for nuclear and mitochondrial DNA, and as a modulator of other transcription factors. Affects inflammatory responses, cellular proliferation and differentiation in target tissues. Could act as a coactivator for STAT5-dependent transcription upon growth hormone (GH) stimulation and could reveal an essential role of hepatic GR in the control of body growth. Involved in chromatin remodeling. May play a negative role in adipogenesis through the regulation of lipolytic and antilipogenic genes expression.
- Gene Name:
- NR3C1
- Uniprot ID:
- P04150
- Molecular Weight:
- 85658.57 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 3.49 uM | NVS_NR_hGR | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. This enzyme contributes to the wide pharmacokinetics variability of the metabolism of drugs such as S-warfarin, diclofenac, phenytoin, tolbutamide and losartan.
- Gene Name:
- CYP2C9
- Uniprot ID:
- P11712
- Molecular Weight:
- 55627.365 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 3.55 uM | NVS_ADME_hCYP2C9 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Nucleoside transmembrane transporter activity
- Specific Function:
- Mediates both influx and efflux of nucleosides across the membrane (equilibrative transporter). It is sensitive (ES) to low concentrations of the inhibitor nitrobenzylmercaptopurine riboside (NBMPR) and is sodium-independent. It has a higher affinity for adenosine. Inhibited by dipyridamole and dilazep (anticancer chemotherapeutics drugs).
- Gene Name:
- SLC29A1
- Uniprot ID:
- Q99808
- Molecular Weight:
- 50218.805 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.01 uM | NVS_TR_hAdoT | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Identical protein binding
- Specific Function:
- Receptor for adenosine. The activity of this receptor is mediated by G proteins which activate adenylyl cyclase.
- Gene Name:
- ADORA2A
- Uniprot ID:
- P29274
- Molecular Weight:
- 44706.925 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.16 uM | NVS_GPCR_hAdoRA2a | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Guanyl-nucleotide exchange factor activity
- Specific Function:
- The muscarinic acetylcholine receptor mediates various cellular responses, including inhibition of adenylate cyclase, breakdown of phosphoinositides and modulation of potassium channels through the action of G proteins. Primary transducing effect is inhibition of adenylate cyclase.
- Gene Name:
- CHRM4
- Uniprot ID:
- P08173
- Molecular Weight:
- 53048.65 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.50 uM | NVS_GPCR_hM4 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.00 uM | NVS_ADME_hCYP2B6 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor activity
- Specific Function:
- Receptor for neuropeptide Y and peptide YY. The rank order of affinity of this receptor for pancreatic polypeptides is PYY > NPY > PYY (3-36) > NPY (2-36) > [Ile-31, Gln-34] PP > [Leu-31, Pro-34] NPY > PP, [Pro-34] PYY and NPY free acid.
- Gene Name:
- NPY2R
- Uniprot ID:
- P49146
- Molecular Weight:
- 42730.69 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.83 uM | NVS_GPCR_hNPY2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein heterodimerization activity
- Specific Function:
- Receptor for angiotensin II. Mediates its action by association with G proteins that activate a phosphatidylinositol-calcium second messenger system.
- Gene Name:
- AGTR1
- Uniprot ID:
- P30556
- Molecular Weight:
- 41060.53 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.83 uM | NVS_GPCR_hAT1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii distal enhancer sequence-specific binding
- Specific Function:
- Transcription activator that binds to antioxidant response (ARE) elements in the promoter regions of target genes. Important for the coordinated up-regulation of genes in response to oxidative stress. May be involved in the transcriptional activation of genes of the beta-globin cluster by mediating enhancer activity of hypersensitive site 2 of the beta-globin locus control region.
- Gene Name:
- NFE2L2
- Uniprot ID:
- Q16236
- Molecular Weight:
- 67825.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.91 uM | ATG_NRF2_ARE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]