Rotenone (T3D0800)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-06-02 15:21:19 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:22:50 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D0800 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Rotenone | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Rotenone is found in jicama. Rotenone is widely distributed in the Leguminosae (Papilionoideae) e.g. Pachyrrhizus erosus (yam bean).Rotenone is an odorless chemical that is used as a broad-spectrum insecticide, piscicide, and pesticide. It occurs naturally in the roots and stems of several plants such as the jicama vine plant. In mammals, including humans, it is linked to the development of Parkinson's disease. (Wikipedia) Rotenone has been shown to exhibit apoptotic, neuroprotectant and neuroprotective functions (1, 2, 2). Rotenone belongs to the family of Rotenoids. These are phenolic compounds containing aA cis-fused tetrahydrochromeno[3,4-b]chromenenucleus. Many rotenoids contain an additional ring, e.g rotenone[1]. (Reference: [1] IUPAC. Compendium of Chemical Terminology, 2nd ed. (the Gold Book). Compiled by A. D. McNaught and A. Wilkinson. Blackwell Scientific Publications, Oxford (1997). XML on-line corrected version: http://goldbook.iupac.org (2006-) created by M. Nic, J. Jirat, B. Kosata; updates compiled by A. Jenkins. ISBN 0-9678550-9-8. doi:10.1351/goldbook. (PAC, 1995, 67, 1307 (Glossary of class names of organic compounds and reactivity intermediates based on structure (IUPAC Recommendations 1995)) on page 1364)). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
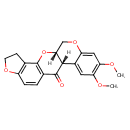
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C20H18O6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 354.353 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 354.110 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 83-79-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | (1S,13S)-16,17-dimethoxy-2,7,20-trioxapentacyclo[11.8.0.0³,¹¹.0⁴,⁸.0¹⁴,¹⁹]henicosa-3(11),4(8),9,14,16,18-hexaen-12-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | (1S,13S)-16,17-dimethoxy-2,7,20-trioxapentacyclo[11.8.0.0³,¹¹.0⁴,⁸.0¹⁴,¹⁹]henicosa-3(11),4(8),9,14,16,18-hexaen-12-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@@]12COC3=CC(OC)=C(OC)C=C3[C@]1([H])C(=O)C1=C(O2)C2=C(OCC2)C=C1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C20H18O6/c1-22-15-7-12-14(8-16(15)23-2)25-9-17-18(12)19(21)11-3-4-13-10(5-6-24-13)20(11)26-17/h3-4,7-8,17-18H,5-6,9H2,1-2H3/t17-,18+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=NZVPMEQJVQAGNB-MSOLQXFVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as rotenones. These are rotenoids with a structure based on a 6a,12a-dihydrochromeno[3,4-b]chromen-12(6H)-one skeleton. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Phenylpropanoids and polyketides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Isoflavonoids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Rotenoids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Rotenones | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Rotenone works by interfering with the electron transport chain in mitochondria. Specifically, it inhibits the transfer of electrons from iron-sulfur centers in complex I to ubiquinone. This prevents NADH from being converted into usable cellular energy (ATP). (5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Rotenone is used in solution as a pesticide and insecticide. It is commonly used in powdered or emulsified liquid form in fisheries management to remove unwanted fish species, such as the eradication of exotic fish from non-native habitats. People catch fish by extracting rotenone from plants and releasing it into water. Poisoned fish come to the surface and are easily caught. This method was first practiced by various indigenous tribes who smashed the roots. Fish caught this way can be eaten because rotenone is very poorly absorbed by the gastrointestinal tract of humans, whereas it is lethal to fish because it readily enters the blood stream of the fish through the gills. Small-scale sampling with rotenone is used by fish researchers studying the biodiversity of marine fishes to collect cryptic, or hidden, fishes, which represent an important component of shoreline fish communities. Rotenone is the most effective tool available because only small quantities are necessary. It has only minor and transient environmental side-effects. Rotenone is also used in powdered form to reduce parasitic mites on chickens and other fowl. In the United States and in Canada, all uses of Rotenone except as a piscicide are being phased out. (5) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Studies with primary cultures of rat neurons and microglia have shown low doses of rotenone (below 10 nM) to induce oxidative damage and death of dopaminergic neurons and it is these neurons in the substantia nigra that die in Parkinson's disease. Injecting rotenone into rats causes symptoms of Parkinson's disease to develop. The study does not directly suggest that rotenone exposure is responsible for Parkinson's disease in humans. (5) Rotenone dust is highly irritating to the eyes (causing conjunctivitis), skin (causing contact dermatitis), upper respiratory tract (causing rhinitis), and throat (linked with pharyngitis) (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Acute poisoning in animals is characterized by an initial respiratory stimulation followed by respiratory depression, ataxia, convulsions, and death from respiratory arrest. (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Since rotenone is unstable in light and heat, its toxicity can be lost after 2 or 3 days. (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB34436 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 6758 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 6500 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C07593 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 28201 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | CPD-5741 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | D012402 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Rotenone | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | 1279 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Rotenone | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D0800.pdf | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Nadh dehydrogenase (ubiquinone) activity
- Specific Function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
- Gene Name:
- MT-ND4
- Uniprot ID:
- P03905
- Molecular Weight:
- 51580.26 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 0.0035 uM | Not Available | BindingDB 50135527 |
References
- General Function:
- Nadh dehydrogenase (ubiquinone) activity
- Specific Function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity). This is the largest subunit of complex I and it is a component of the iron-sulfur (IP) fragment of the enzyme. It may form part of the active site crevice where NADH is oxidized.
- Gene Name:
- NDUFS1
- Uniprot ID:
- P28331
- Molecular Weight:
- 79466.77 Da
References
- Wikipedia. Rotenone. Last Updated 21 July 2009. [Link]
- General Function:
- Nadh dehydrogenase (ubiquinone) activity
- Specific Function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
- Gene Name:
- MT-ND1
- Uniprot ID:
- P03886
- Molecular Weight:
- 35660.055 Da
References
- Wikipedia. Rotenone. Last Updated 21 July 2009. [Link]
- General Function:
- Nadh dehydrogenase (ubiquinone) activity
- Specific Function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
- Gene Name:
- MT-ND2
- Uniprot ID:
- P03891
- Molecular Weight:
- 38960.47 Da
References
- Wikipedia. Rotenone. Last Updated 21 July 2009. [Link]
- General Function:
- Nadh dehydrogenase (ubiquinone) activity
- Specific Function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
- Gene Name:
- MT-ND3
- Uniprot ID:
- P03897
- Molecular Weight:
- 13185.87 Da
References
- Wikipedia. Rotenone. Last Updated 21 July 2009. [Link]
- General Function:
- Temperature-gated cation channel activity
- Specific Function:
- Receptor-activated non-selective cation channel involved in detection of pain and possibly also in cold perception and inner ear function (PubMed:25389312, PubMed:25855297). Has a central role in the pain response to endogenous inflammatory mediators and to a diverse array of volatile irritants, such as mustard oil, cinnamaldehyde, garlic and acrolein, an irritant from tears gas and vehicule exhaust fumes (PubMed:25389312, PubMed:20547126). Is also activated by menthol (in vitro)(PubMed:25389312). Acts also as a ionotropic cannabinoid receptor by being activated by delta(9)-tetrahydrocannabinol (THC), the psychoactive component of marijuana (PubMed:25389312). May be a component for the mechanosensitive transduction channel of hair cells in inner ear, thereby participating in the perception of sounds. Probably operated by a phosphatidylinositol second messenger system (By similarity).
- Gene Name:
- TRPA1
- Uniprot ID:
- O75762
- Molecular Weight:
- 127499.88 Da
References
- Nilius B, Prenen J, Owsianik G: Irritating channels: the case of TRPA1. J Physiol. 2011 Apr 1;589(Pt 7):1543-9. doi: 10.1113/jphysiol.2010.200717. Epub 2010 Nov 15. [21078588 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1.
- Gene Name:
- ESR1
- Uniprot ID:
- P03372
- Molecular Weight:
- 66215.45 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.00 uM | ACEA_T47D_80hr_Positive | ACEA Biosciences |
| AC50 | 0.04 uM | Tox21_ERa_BLA_Agonist_ratio | Tox21/NCGC |
| AC50 | 0.02 uM | Tox21_ERa_LUC_BG1_Antagonist | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii distal enhancer sequence-specific binding
- Specific Function:
- Transcription activator that binds to antioxidant response (ARE) elements in the promoter regions of target genes. Important for the coordinated up-regulation of genes in response to oxidative stress. May be involved in the transcriptional activation of genes of the beta-globin cluster by mediating enhancer activity of hypersensitive site 2 of the beta-globin locus control region.
- Gene Name:
- NFE2L2
- Uniprot ID:
- Q16236
- Molecular Weight:
- 67825.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.01 uM | ATG_NRF2_ARE_CIS | Attagene |
| AC50 | 0.01 uM | ATG_NRF2_ARE_CIS | Attagene |
| AC50 | 0.01 uM | ATG_NRF2_ARE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Steroid hormone receptors are ligand-activated transcription factors that regulate eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Transcription factor activity is modulated by bound coactivator and corepressor proteins. Transcription activation is down-regulated by NR0B2. Activated, but not phosphorylated, by HIPK3 and ZIPK/DAPK3.
- Gene Name:
- AR
- Uniprot ID:
- P10275
- Molecular Weight:
- 98987.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.02 uM | Tox21_AR_LUC_MDAKB2_Antagonist | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor signaling protein serine/threonine kinase activity
- Specific Function:
- On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. Binds to BMP-7, BMP-2 and, less efficiently, BMP-4. Binding is weak but enhanced by the presence of type I receptors for BMPs.
- Gene Name:
- BMPR2
- Uniprot ID:
- Q13873
- Molecular Weight:
- 115200.475 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.03 uM | ATG_BRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcription regulatory region sequence-specific dna binding
- Specific Function:
- Transcription factor that binds to the octamer motif (5'-ATTTGCAT-3') and activates the promoters of the genes for some small nuclear RNAs (snRNA) and of genes such as those for histone H2B and immunoglobulins. Modulates transcription transactivation by NR3C1, AR and PGR (By similarity). In case of human herpes simplex virus (HSV) infection, POU2F1 forms a multiprotein-DNA complex with the viral transactivator protein VP16 and HCFC1 thereby enabling the transcription of the viral immediate early genes.
- Gene Name:
- POU2F1
- Uniprot ID:
- P14859
- Molecular Weight:
- 76470.82 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.03 uM | ATG_Oct_MLP_CIS | Attagene |
| AC50 | 0.03 uM | ATG_Oct_MLP_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii core promoter proximal region sequence-specific binding
- Specific Function:
- Endoplasmic reticulum (ER)-bound transcription factor that plays a role in the unfolded protein response (UPR). Involved in cell proliferation and migration, tumor suppression and inflammatory gene expression. Plays also a role in the human immunodeficiency virus type 1 (HIV-1) virus protein expression and in the herpes simplex virus-1 (HSV-1) latent infection and reactivation from latency. Isoform 2 plays a role in the unfolded protein response (UPR). Isoform 2 acts as a positive regulator of LKN-1/CCL15-induced chemotaxis signaling of leukocyte cell migration. Isoform 2 may play a role as a cellular tumor suppressor that is targeted by the hepatitis C virus (HSV) core protein. Isoform 2 represses the VP16-mediated transactivation of immediate early genes of the HSV-1 virus by sequestring host cell factor-1 HCFC1 in the ER membrane of sensory neurons, thereby preventing the initiation of the replicative cascade leading to latent infection. Isoform 3 functions as a negative transcriptional regulator in ligand-induced transcriptional activation of the glucocorticoid receptor NR3C1 by recruiting and activating histone deacetylases (HDAC1, HDAC2 and HDAC6). Isoform 3 decreases the acetylation level of histone H4. Isoform 3 does not promote the chemotactic activity of leukocyte cells.Processed cyclic AMP-responsive element-binding protein 3: acts as a transcription factor that activates unfolded protein response (UPR) target genes during endoplasmic reticulum (ER) stress response. Promotes cell survival against ER stress-induced apoptotic cell death during UPR. Activates transcription from CRE and C/EBP-containing reporter genes. Induces transcriptional activation of chemokine receptors. Activates transcription of genes required for reactivation of the latent HSV-1 virus. Down-regulates Tat-dependent transcription of the HIV-1 LTR by interacting with HIV-1 Tat. It's transcriptional activity is inhibited by CREBZF in a HCFC1-dependent manner, by the viral transactivator protein VP16 and by the HCV core protein. Binds DNA to the cAMP response element (CRE) (consensus: 5'-GTGACGT[AG][AG]-3') and C/EBP sequences present in many viral and cellular promoters. Binds to the unfolded protein respons element (UPRE) consensus sequences sites. Binds DNA to the 5'-CCAC[GA]-3'half of ERSE II (5'-ATTGG-N-CCACG-3'). Associates with chromatin to the HERPUD1 promoter.
- Gene Name:
- CREB3
- Uniprot ID:
- O43889
- Molecular Weight:
- 43916.65 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.07 uM | ATG_CRE_CIS | Attagene |
| AC50 | 0.07 uM | ATG_CRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety. Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. May be required for the propagation of clock information to metabolic pathways regulated by PER2.
- Gene Name:
- PPARA
- Uniprot ID:
- Q07869
- Molecular Weight:
- 52224.595 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.07 uM | ATG_PPRE_CIS | Attagene |
| AC50 | 0.07 uM | ATG_PPRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Iron ion binding
- Specific Function:
- Catalyzes the conversion of 25-hydroxyvitamin D3 (25(OH)D) to 1-alpha,25-dihydroxyvitamin D3 (1,25(OH)2D) plays an important role in normal bone growth, calcium metabolism, and tissue differentiation.
- Gene Name:
- CYP27B1
- Uniprot ID:
- O15528
- Molecular Weight:
- 56503.475 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.08 uM | ATG_VDRE_CIS | Attagene |
| AC50 | 0.08 uM | ATG_VDRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii core promoter proximal region sequence-specific binding
- Specific Function:
- Activates the metallothionein I promoter. Binds to the metal responsive element (MRE).
- Gene Name:
- MTF1
- Uniprot ID:
- Q14872
- Molecular Weight:
- 80956.22 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.09 uM | ATG_MRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Polycomb group (PcG) that specifically binds histone H3 trimethylated at 'Lys-36' (H3K36me3) and recruits the PRC2 complex. Acts by binding to H3K36me3, a mark for transcriptional activation, and recruiting the PRC2 complex, leading to enhance PRC2 H3K27me3 methylation activity. Regulates the transcriptional networks during embryonic stem cell self-renewal and differentiation. Promotes recruitment of the PRC2 complex to the inactive X chromosome in differentiating XX ES cells and PRC2 recruitment to target genes in undifferentiated ES cells. Required to repress Hox genes by enhancing H3K27me3 methylation of the PRC2 complex. In some conditions may act as an inhibitor of PRC2 activity: able to activate the CDKN2A gene and promote cellular senescence by suppressing the catalytic activity of the PRC2 complex locally. Binds to the metal-regulating-element (MRE) of MT1A gene promoter (By similarity).
- Gene Name:
- MTF2
- Uniprot ID:
- Q9Y483
- Molecular Weight:
- 67105.08 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.10 uM | ATG_MRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds DNA as a monomer to ROR response elements (RORE) containing a single core motif half-site 5'-AGGTCA-3' preceded by a short A-T-rich sequence. Key regulator of embryonic development, cellular differentiation, immunity, circadian rhythm as well as lipid, steroid, xenobiotics and glucose metabolism. Considered to have intrinsic transcriptional activity, have some natural ligands like oxysterols that act as agonists (25-hydroxycholesterol) or inverse agonists (7-oxygenated sterols), enhancing or repressing the transcriptional activity, respectively. Recruits distinct combinations of cofactors to target genes regulatory regions to modulate their transcriptional expression, depending on the tissue, time and promoter contexts. Regulates genes involved in photoreceptor development including OPN1SW, OPN1SM and ARR3 and skeletal muscle development with MYOD1. Required for proper cerebellum development, regulates SHH gene expression, among others, to induce granule cells proliferation as well as expression of genes involved in calcium-mediated signal transduction. Regulates the circadian expression of several clock genes, including CLOCK, ARNTL/BMAL1, NPAS2 and CRY1. Competes with NR1D1 for binding to their shared DNA response element on some clock genes such as ARNTL/BMAL1, CRY1 and NR1D1 itself, resulting in NR1D1-mediated repression or RORA-mediated activation of clock genes expression, leading to the circadian pattern of clock genes expression. Therefore influences the period length and stability of the clock. Regulates genes involved in lipid metabolism such as apolipoproteins APOA1, APOA5, APOC3 and PPARG. In liver, has specific and redundant functions with RORC as positive or negative modulator of expression of genes encoding phase I and phase II proteins involved in the metabolism of lipids, steroids and xenobiotics, such as CYP7B1 and SULT2A1. Induces a rhythmic expression of some of these genes. In addition, interplays functionally with NR1H2 and NR1H3 for the regulation of genes involved in cholesterol metabolism. Also involved in the regulation of hepatic glucose metabolism through the modulation of G6PC and PCK1. In adipose tissue, plays a role as negative regulator of adipocyte differentiation, probably acting through dual mechanisms. May suppress CEBPB-dependent adipogenesis through direct interaction and PPARG-dependent adipogenesis through competition for DNA-binding. Downstream of IL6 and TGFB and synergistically with RORC isoform 2, is implicated in the lineage specification of uncommitted CD4(+) T-helper (T(H)) cells into T(H)17 cells, antagonizing the T(H)1 program. Probably regulates IL17 and IL17F expression on T(H) by binding to the essential enhancer conserved non-coding sequence 2 (CNS2) in the IL17-IL17F locus. Involved in hypoxia signaling by interacting with and activating the transcriptional activity of HIF1A. May inhibit cell growth in response to cellular stress. May exert an anti-inflammatory role by inducing CHUK expression and inhibiting NF-kappa-B signaling.
- Gene Name:
- RORA
- Uniprot ID:
- P35398
- Molecular Weight:
- 58974.35 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.19 uM | ATG_RORE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Binds and transactivates the retinoic acid response elements that control expression of the retinoic acid receptor beta 2 and alcohol dehydrogenase 3 genes. Transactivates both the phenobarbital responsive element module of the human CYP2B6 gene and the CYP3A4 xenobiotic response element.
- Gene Name:
- NR1I3
- Uniprot ID:
- Q14994
- Molecular Weight:
- 39942.145 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.23 uM | ATG_PBREM_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Responsible for the metabolism of a number of therapeutic agents such as the anticonvulsant drug S-mephenytoin, omeprazole, proguanil, certain barbiturates, diazepam, propranolol, citalopram and imipramine.
- Gene Name:
- CYP2C19
- Uniprot ID:
- P33261
- Molecular Weight:
- 55930.545 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.51 uM | NVS_ADME_hCYP2C19 | Novascreen |
| AC50 | 0.42 uM | NVS_ADME_hCYP2C19 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxygen binding
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP3A5
- Uniprot ID:
- P20815
- Molecular Weight:
- 57108.065 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.20 uM | NVS_ADME_hCYP3A5 | Novascreen |
| AC50 | 0.46 uM | NVS_ADME_hCYP3A5 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- High-capacity urate exporter functioning in both renal and extrarenal urate excretion. Plays a role in porphyrin homeostasis as it is able to mediates the export of protoporhyrin IX (PPIX) both from mitochondria to cytosol and from cytosol to extracellular space, and cellular export of hemin, and heme. Xenobiotic transporter that may play an important role in the exclusion of xenobiotics from the brain. Appears to play a major role in the multidrug resistance phenotype of several cancer cell lines. Implicated in the efflux of numerous drugs and xenobiotics: mitoxantrone, the photosensitizer pheophorbide, camptothecin, methotrexate, azidothymidine (AZT), and the anthracyclines daunorubicin and doxorubicin.
- Gene Name:
- ABCG2
- Uniprot ID:
- Q9UNQ0
- Molecular Weight:
- 72313.47 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.58 uM | CLZD_ABCG2_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Glutathione transferase activity
- Specific Function:
- Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles.
- Gene Name:
- GSTA2
- Uniprot ID:
- P09210
- Molecular Weight:
- 25663.675 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 2.83 uM | CLZD_GSTA2_24 | CellzDirect |
| AC50 | 0.63 uM | CLZD_GSTA2_48 | CellzDirect |
| AC50 | 0.63 uM | CLZD_GSTA2_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxygen binding
- Specific Function:
- Catalyzes the formation of aromatic C18 estrogens from C19 androgens.
- Gene Name:
- CYP19A1
- Uniprot ID:
- P11511
- Molecular Weight:
- 57882.48 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.67 uM | Tox21_Aromatase_Inhibition | Tox21/NCGC |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d 24-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP1A1
- Uniprot ID:
- P04798
- Molecular Weight:
- 58164.815 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.00 uM | CLZD_CYP1A1_24 | CellzDirect |
| AC50 | 0.70 uM | CLZD_CYP1A1_48 | CellzDirect |
| AC50 | 0.70 uM | CLZD_CYP1A1_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Isoform Alpha-1: Nuclear hormone receptor that can act as a repressor or activator of transcription. High affinity receptor for thyroid hormones, including triiodothyronine and thyroxine.Isoform Alpha-2: Does not bind thyroid hormone and functions as a weak dominant negative inhibitor of thyroid hormone action.
- Gene Name:
- THRA
- Uniprot ID:
- P10827
- Molecular Weight:
- 54815.055 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.88 uM | ATG_THRa1_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins that stimulate adenylate cyclase. It has a high affinity for tricyclic psychotropic drugs (By similarity). Controls pyramidal neurons migration during corticogenesis, through the regulation of CDK5 activity (By similarity). Is an activator of TOR signaling (PubMed:23027611).
- Gene Name:
- HTR6
- Uniprot ID:
- P50406
- Molecular Weight:
- 46953.625 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.24 uM | NVS_GPCR_h5HT6 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transferase activity
- Specific Function:
- Synthesizes the second messagers cyclic ADP-ribose and nicotinate-adenine dinucleotide phosphate, the former a second messenger for glucose-induced insulin secretion. Also has cADPr hydrolase activity. Also moonlights as a receptor in cells of the immune system.
- Gene Name:
- CD38
- Uniprot ID:
- P28907
- Molecular Weight:
- 34328.145 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SAg_CD38_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- CCL2
- Uniprot ID:
- P13500
- Molecular Weight:
- 11024.87 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_LPS_MCP1_down | BioSeek |
| AC50 | 1.48 uM | BSK_SAg_MCP1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Cell-surface glycoprotein having a role in immunoadhesion. Mediates in the adhesion of blood neutrophils in cytokine-activated endothelium through interaction with PSGL1/SELPLG. May have a role in capillary morphogenesis.
- Gene Name:
- SELE
- Uniprot ID:
- P16581
- Molecular Weight:
- 66654.575 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_LPS_Eselectin_down | BioSeek |
| AC50 | 1.48 uM | BSK_SAg_Eselectin_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Involved in lymphocyte proliferation and functions as a signal transmitting receptor in lymphocytes, natural killer (NK) cells, and platelets.
- Gene Name:
- CD69
- Uniprot ID:
- Q07108
- Molecular Weight:
- 22559.25 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SAg_CD69_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses. Known ligands include EGF, TGFA/TGF-alpha, amphiregulin, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF. Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules. May also activate the NF-kappa-B signaling cascade. Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling. Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin.Isoform 2 may act as an antagonist of EGF action.
- Gene Name:
- EGFR
- Uniprot ID:
- P00533
- Molecular Weight:
- 134276.185 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_EGFR_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Peptide antigen binding
- Specific Function:
- Binds peptides derived from antigens that access the endocytic route of antigen presenting cells (APC) and presents them on the cell surface for recognition by the CD4 T-cells. The peptide binding cleft accommodates peptides of 10-30 residues. The peptides presented by MHC class II molecules are generated mostly by degradation of proteins that access the endocytic route, where they are processed by lysosomal proteases and other hydrolases. Exogenous antigens that have been endocytosed by the APC are thus readily available for presentation via MHC II molecules, and for this reason this antigen presentation pathway is usually referred to as exogenous. As membrane proteins on their way to degradation in lysosomes as part of their normal turn-over are also contained in the endosomal/lysosomal compartments, exogenous antigens must compete with those derived from endogenous components. Autophagy is also a source of endogenous peptides, autophagosomes constitutively fuse with MHC class II loading compartments. In addition to APCs, other cells of the gastrointestinal tract, such as epithelial cells, express MHC class II molecules and CD74 and act as APCs, which is an unusual trait of the GI tract. To produce a MHC class II molecule that presents an antigen, three MHC class II molecules (heterodimers of an alpha and a beta chain) associate with a CD74 trimer in the ER to form a heterononamer. Soon after the entry of this complex into the endosomal/lysosomal system where antigen processing occurs, CD74 undergoes a sequential degradation by various proteases, including CTSS and CTSL, leaving a small fragment termed CLIP (class-II-associated invariant chain peptide). The removal of CLIP is facilitated by HLA-DM via direct binding to the alpha-beta-CLIP complex so that CLIP is released. HLA-DM stabilizes MHC class II molecules until primary high affinity antigenic peptides are bound. The MHC II molecule bound to a peptide is then transported to the cell membrane surface. In B-cells, the interaction between HLA-DM and MHC class II molecules is regulated by HLA-DO. Primary dendritic cells (DCs) also to express HLA-DO. Lysosomal microenvironment has been implicated in the regulation of antigen loading into MHC II molecules, increased acidification produces increased proteolysis and efficient peptide loading.
- Gene Name:
- HLA-DRA
- Uniprot ID:
- P01903
- Molecular Weight:
- 28606.685 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_BE3C_hLADR_down | BioSeek |
| AC50 | 1.48 uM | BSK_3C_hLADR_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Interleukin-8 receptor binding
- Specific Function:
- IL-8 is a chemotactic factor that attracts neutrophils, basophils, and T-cells, but not monocytes. It is also involved in neutrophil activation. It is released from several cell types in response to an inflammatory stimulus. IL-8(6-77) has a 5-10-fold higher activity on neutrophil activation, IL-8(5-77) has increased activity on neutrophil activation and IL-8(7-77) has a higher affinity to receptors CXCR1 and CXCR2 as compared to IL-8(1-77), respectively.
- Gene Name:
- CXCL8
- Uniprot ID:
- P10145
- Molecular Weight:
- 11097.98 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SAg_IL8_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Prostaglandin e receptor activity
- Specific Function:
- Receptor for prostaglandin E2 (PGE2). The activity of this receptor is mediated by G(s) proteins that stimulate adenylate cyclase. The subsequent raise in intracellular cAMP is responsible for the relaxing effect of this receptor on smooth muscle.
- Gene Name:
- PTGER2
- Uniprot ID:
- P43116
- Molecular Weight:
- 39759.945 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_LPS_PGE2_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Tumor necrosis factor receptor binding
- Specific Function:
- Cytokine that binds to TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. It is mainly secreted by macrophages and can induce cell death of certain tumor cell lines. It is potent pyrogen causing fever by direct action or by stimulation of interleukin-1 secretion and is implicated in the induction of cachexia, Under certain conditions it can stimulate cell proliferation and induce cell differentiation. Impairs regulatory T-cells (Treg) function in individuals with rheumatoid arthritis via FOXP3 dephosphorylation. Upregulates the expression of protein phosphatase 1 (PP1), which dephosphorylates the key 'Ser-418' residue of FOXP3, thereby inactivating FOXP3 and rendering Treg cells functionally defective (PubMed:23396208). Key mediator of cell death in the anticancer action of BCG-stimulated neutrophils in combination with DIABLO/SMAC mimetic in the RT4v6 bladder cancer cell line (PubMed:22517918).The TNF intracellular domain (ICD) form induces IL12 production in dendritic cells.
- Gene Name:
- TNF
- Uniprot ID:
- P01375
- Molecular Weight:
- 25644.15 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_LPS_TNFa_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Receptor for TNFSF5/CD40LG. Transduces TRAF6- and MAP3K8-mediated signals that activate ERK in macrophages and B cells, leading to induction of immunoglobulin secretion.
- Gene Name:
- CD40
- Uniprot ID:
- P25942
- Molecular Weight:
- 30618.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_LPS_CD40_down | BioSeek |
| AC50 | 1.48 uM | BSK_SAg_CD40_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Urokinase plasminogen activator receptor activity
- Specific Function:
- Acts as a receptor for urokinase plasminogen activator. Plays a role in localizing and promoting plasmin formation. Mediates the proteolysis-independent signal transduction activation effects of U-PA. It is subject to negative-feedback regulation by U-PA which cleaves it into an inactive form.
- Gene Name:
- PLAUR
- Uniprot ID:
- Q03405
- Molecular Weight:
- 36977.62 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_3C_uPAR_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serine-type endopeptidase activity
- Specific Function:
- Specifically cleaves the zymogen plasminogen to form the active enzyme plasmin.
- Gene Name:
- PLAU
- Uniprot ID:
- P00749
- Molecular Weight:
- 48507.09 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_BE3C_uPA_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Primary amine oxidase activity
- Specific Function:
- Important in cell-cell recognition. Appears to function in leukocyte-endothelial cell adhesion. Interacts with integrin alpha-4/beta-1 (ITGA4/ITGB1) on leukocytes, and mediates both adhesion and signal transduction. The VCAM1/ITGA4/ITGB1 interaction may play a pathophysiologic role both in immune responses and in leukocyte emigration to sites of inflammation.
- Gene Name:
- VCAM1
- Uniprot ID:
- P19320
- Molecular Weight:
- 81275.43 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_VCAM1_down | BioSeek |
| AC50 | 1.48 uM | BSK_LPS_VCAM1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.50 uM | CLZD_CYP3A4_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transporter activity
- Specific Function:
- Involved in the ATP-dependent secretion of bile salts into the canaliculus of hepatocytes.
- Gene Name:
- ABCB11
- Uniprot ID:
- O95342
- Molecular Weight:
- 146405.83 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 6.07 uM | CLZD_ABCB11_24 | CellzDirect |
| AC50 | 1.57 uM | CLZD_ABCB11_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.82 uM | CLZD_CYP2B6_24 | CellzDirect |
| AC50 | 1.63 uM | CLZD_CYP2B6_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Sulfotransferase activity
- Specific Function:
- Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) as sulfonate donor to catalyze the sulfonation of steroids and bile acids in the liver and adrenal glands.
- Gene Name:
- SULT2A1
- Uniprot ID:
- Q06520
- Molecular Weight:
- 33779.57 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.58 uM | CLZD_SULT2A1_24 | CellzDirect |
| AC50 | 1.69 uM | CLZD_SULT2A1_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. This enzyme contributes to the wide pharmacokinetics variability of the metabolism of drugs such as S-warfarin, diclofenac, phenytoin, tolbutamide and losartan.
- Gene Name:
- CYP2C9
- Uniprot ID:
- P11712
- Molecular Weight:
- 55627.365 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 3.75 uM | CLZD_CYP2C9_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protease binding
- Specific Function:
- Metalloproteinase inhibitor that functions by forming one to one complexes with target metalloproteinases, such as collagenases, and irreversibly inactivates them by binding to their catalytic zinc cofactor. Acts on MMP1, MMP2, MMP3, MMP7, MMP8, MMP9, MMP10, MMP11, MMP12, MMP13 and MMP16. Does not act on MMP14. Also functions as a growth factor that regulates cell differentiation, migration and cell death and activates cellular signaling cascades via CD63 and ITGB1. Plays a role in integrin signaling. Mediates erythropoiesis in vitro; but, unlike IL3, it is species-specific, stimulating the growth and differentiation of only human and murine erythroid progenitors.
- Gene Name:
- TIMP1
- Uniprot ID:
- P01033
- Molecular Weight:
- 23170.64 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_TIMP1_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. The high affinity ligand for RXRs is 9-cis retinoic acid. RXRA serves as a common heterodimeric partner for a number of nuclear receptors. The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence of ligand, the RXR-RAR heterodimers associate with a multiprotein complex containing transcription corepressors that induce histone acetylation, chromatin condensation and transcriptional suppression. On ligand binding, the corepressors dissociate from the receptors and associate with the coactivators leading to transcriptional activation. The RXRA/PPARA heterodimer is required for PPARA transcriptional activity on fatty acid oxidation genes such as ACOX1 and the P450 system genes.
- Gene Name:
- RXRA
- Uniprot ID:
- P19793
- Molecular Weight:
- 50810.835 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.12 uM | OT_NURR1_NURR1RXRa_0480 | Odyssey Thera |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- TP53
- Uniprot ID:
- P04637
- Molecular Weight:
- 43652.79 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.02 uM | APR_p53Act_72h_up | Apredica |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]