| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-06-18 21:54:33 UTC |
|---|
| Update Date | 2014-12-24 20:23:07 UTC |
|---|
| Accession Number | T3D1088 |
|---|
| Identification |
|---|
| Common Name | (±)-Metalaxyl |
|---|
| Class | Small Molecule |
|---|
| Description | (±)-Metalaxyl is a systemic agricultural fungicide belonging to the family of Depsipeptides. These are natural or synthetic compounds having sequences of amino and hydroxy carboxylic acid residues (usually I-amino and I-hydroxy acids), commonly but not necessarily regularly alternating. |
|---|
| Compound Type | - Amide
- Amine
- Aromatic Hydrocarbon
- Chloroacetanilide
- Ester
- Ether
- Food Toxin
- Household Toxin
- Metabolite
- Organic Compound
- Organochloride
- Pesticide
- Synthetic Compound
|
|---|
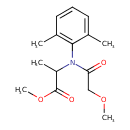
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (+-)-Metalaxyl | | (r)-Metalaxyl | | (±)-metalaxyl | | Allegiance | | Apron | | Apron 2E | | Apron FL | | Apron SD 35 | | Caswell No. 375AA | | D,L-N-(2,6-Dimethylphenyl)-N-(2'-methoxyacetyl)alaninate de methyle | | Mefenoxam | | Metalasyl | | Metalaxil | | Metalaxyl | | Metanaxin | | Metasyl | | Metaxanin | | Methyl 2-[(methoxyacetyl)-2,6-dimethylanilino]propanoate | | Methyl N-(2,6-dimethylphenyl)-N-(methoxyacetyl)-DL-alaninate | | Methyl N-(2-methoxyacetyl)-N-(2,6-xylyl)-DL-alaninate | | N-(2,6-Dimethylphenyl)-N-(methoxyacetyl)-alanine methyl ester | | rac-Metalaxyl | | Ridomil 2E | | Ridomil 72WP | | Ridomil vino | | Subdue 2E | | Subdue 5SP |
|
|---|
| Chemical Formula | C15H21NO4 |
|---|
| Average Molecular Mass | 279.332 g/mol |
|---|
| Monoisotopic Mass | 279.147 g/mol |
|---|
| CAS Registry Number | 57837-19-1 |
|---|
| IUPAC Name | methyl 2-[N-(2,6-dimethylphenyl)-2-methoxyacetamido]propanoate |
|---|
| Traditional Name | (+-)-metalaxyl |
|---|
| SMILES | COCC(=O)N(C(C)C(=O)OC)C1=C(C)C=CC=C1C |
|---|
| InChI Identifier | InChI=1/C15H21NO4/c1-10-7-6-8-11(2)14(10)16(13(17)9-19-4)12(3)15(18)20-5/h6-8,12H,9H2,1-5H3 |
|---|
| InChI Key | InChIKey=ZQEIXNIJLIKNTD-UHFFFAOYNA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acid esters. These are ester derivatives of alpha amino acids. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acid esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid ester
- Alanine or derivatives
- Anilide
- M-xylene
- Xylene
- Monocyclic benzene moiety
- Benzenoid
- Tertiary carboxylic acid amide
- Methyl ester
- Carboxylic acid ester
- Carboxamide group
- Monocarboxylic acid or derivatives
- Ether
- Dialkyl ether
- Organopnictogen compound
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | Colorless, odorless crystal (8). |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 71 - 72°C | | Boiling Point | Not Available | | Solubility | 8.4 mg/mL at 22°C [TOMLIN,C (1997)] | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0592-3930000000-d56e2942ba3f6e452344 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0002-0090000000-aad829151558e72524f6 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0090000000-2591a5eaf859e460f866 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-006x-0970000000-d0bbdb2f36069fe95a29 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-01ox-0910000000-33ece08e09c98f660242 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03di-0900000000-3e0d4d4098a62b892e27 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03ea-0900000000-e062baf0dcfc758aedc2 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0900000000-e113e4cc9de04128ed78 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0090000000-4534a8177ab4532a8e38 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-006x-0970000000-c891e8d01ae69479d14c | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-01ox-0910000000-0830c1f0a544a6ba9de5 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03di-0900000000-de907346a6e657727d0b | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03ea-0900000000-f0e6fbc8971da01db24a | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0900000000-eaafbbda1ffa7d96c621 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0002-0090000000-82f49b397d4027b60e85 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-022d-0970000000-9799c863c08322bc95a3 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 75V, Positive | splash10-01qa-0900000000-22fa4e788c2d25ab89f7 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 60V, Positive | splash10-03di-0900000000-cae67fcffc1d51ba2b58 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 75V, Positive | splash10-01qa-0900000000-700f91fa7518b38b9437 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0002-0090000000-3efd480f1c939fce059f | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0090000000-6264726ac6e0d497a860 | 2016-08-02 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00ej-4390000000-919b2c2707afbecd452e | 2016-08-02 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-1900000000-e040c95fea0f0da19cbe | 2016-08-02 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004j-0090000000-9fa282d3b264d46721d9 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002b-2290000000-ace0e4668aacb7f19324 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05fu-7920000000-fb750174ac7f551477c1 | 2016-08-03 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0002-9820000000-d8cdd771b6de936d1891 | 2014-09-20 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | Not Available | 2014-09-20 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100.40 MHz, CDCl3, experimental) | Not Available | 2014-09-23 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Inhalation (8) ; oral (8) ; dermal (8) ; eye contact (8). |
|---|
| Mechanism of Toxicity | Binds to nAChRs in nervous systems. It also causes endocrine disruption in humans by binding to and inhibiting the estrogen receptor. (3, 2) |
|---|
| Metabolism | Three major and one minor metabolic pathways are proposed. One pathway involves hydrolysis of the ether, followed by oxidation of the resulting alcohol, ester hydrolysis, or N-dealkylation of the ester chain. A second pathway involves oxidation of an aromatic methyl to the benzylic acid or ester hydrolysis. The third major pathway is ester hydrolysis, sometimes followed by benzylic acid formation. The minor pathway involves hydroxylation at the meta position of the phenyl ring. The majority of urinary metabolites are conjugated (glucuronide or sulfate) whereas fecal metabolites are mostly unconjugated. The major metabolite in urine & feces is N-(2,6-dimethylphenyl)-N-(hydroxyacetyl) alanine. Metalaxyl is excreted in urine and the feces. (9) |
|---|
| Toxicity Values | LD50: 669 mg/kg (Oral, Rat) (9)
LD50: 7120 mg/kg (Oral, hamster) (9)
LD50: >6000 mg/kg (Dermal, Rabbit) (9) |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Widely used selective herbicide worldwide in corn, soybean and other crop cultures. Elevated concentrations of this herbicide and its degradation products have been detected in surface and groundwater. (1) |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | ARDS/acute lung injury, burns of the esophagus or gastrointestinal tract can result from metalaxyl poisoning. (4) |
|---|
| Symptoms | Metalaxyl is mildly irritating to the skin and eyes. Exposure to metalaxyl often results in such nonspecific symptoms as headache, dizziness, weakness, and nausea. Metaxyl poisoning can produce an allergic hypersensitivity dermatitis or asthma with bronchospasm and wheezing with chronic exposure. (4) |
|---|
| Treatment | Consider gastric lavage, as well was dilution with milk or water after ingestion. Administer charcoal as a slurry following ingestion; however, activated charcoal should not be given to patients ingesting strong acidic or basic caustic chemicals. In case of inhalation, move patient to fresh air. Monitor for respiratory distress. If cough or difficulty breathing develops, evaluate for respiratory tract irritation, bronchitis, or pneumonitis. Administer oxygen and assist ventilation as required. Treat bronchospasm with inhaled beta2 agonist and oral or parenteral corticosteroids. Irrigate exposed eyes with copious amounts of room temperature water for at least 15 minutes. Following dermal exposure, remove contaminated clothing and wash exposed area thoroughly with soap and water. Treat dermal irritation or burns with standard topical therapy. Patients developing dermal hypersensitivity reactions may require treatment with systemic or topical corticosteroids or antihistamines. Administer symptomatic treatment as necessary. (4) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB31802 |
|---|
| PubChem Compound ID | 42586 |
|---|
| ChEMBL ID | Not Available |
|---|
| ChemSpider ID | 38839 |
|---|
| KEGG ID | C10947 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 6790 |
|---|
| BioCyc ID | CPD0-1558 |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Metalaxyl |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | 6479 |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D1088.pdf |
|---|
| General References | - Osano O, Admiraal W, Klamer HJ, Pastor D, Bleeker EA: Comparative toxic and genotoxic effects of chloroacetanilides, formamidines and their degradation products on Vibrio fischeri and Chironomus riparius. Environ Pollut. 2002;119(2):195-202. [12152826 ]
- Taccone-Gallucci M, Manca-di-Villahermosa S, Battistini L, Stuffler RG, Tedesco M, Maccarrone M: N-3 PUFAs reduce oxidative stress in ESRD patients on maintenance HD by inhibiting 5-lipoxygenase activity. Kidney Int. 2006 Apr;69(8):1450-4. [16531984 ]
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- Rumack BH (2009). POISINDEX(R) Information System. Englewood, CO: Micromedex, Inc. CCIS Volume 141, edition expires Aug, 2009.
- Luft S, Milki E, Glustrom E, Ampiah-Bonney R, O'Hara P. Binding of Organochloride and Pyrethroid Pesticides To Estrogen Receptors α and β: A Fluorescence Polarization Assay. Biophysical Journal 2009;96(3):444a.
- Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC.
- Wikipedia. Metalaxyl. Last Updated 22 July 2009. [Link]
- Extension Toxicology Network (1996). Pesticide Information Profile for Metalaxyl. A Pesticide Information Project of Cooperative Extension Offices of Cornell University, Michigan State University, Oregon State University, and University of California at Davis. [Link]
- USEPA (2002). Reregistration Eligibility Decision - Metalaxyl. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|