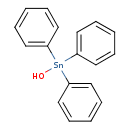
Hydroxytriphenylstannane (T3D1252)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-06-19 21:58:30 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:23:30 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D1252 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Hydroxytriphenylstannane | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Hydroxytriphenylstannane is an Agricultural fungicide for potatoes, sugar beet, pecans and peanuts Hydroxytriphenylstannane belongs to the family of Aromatic Homomonocyclic Compounds. These are aromatic compounds containig only one ring, which is homocyclic. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C18H19OSn | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 370.050 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 371.046 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 76-87-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | triphenylstannanol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | triphenyltin hydroxide | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [OH-].[SnH3+].[C]1=CC=CC=C1.[C]1=CC=CC=C1.[C]1=CC=CC=C1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/3C6H5.H2O.Sn.3H/c3*1-2-4-6-5-3-1;;;;;/h3*1-5H;1H2;;;;/q;;;;+1;;;/p-1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=NEMWKJDHCBHVMD-UHFFFAOYSA-M | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as benzene and substituted derivatives. These are aromatic compounds containing one monocyclic ring system consisting of benzene. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Benzenoids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzene and substituted derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Benzene and substituted derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic homomonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Oral (7) ; inhalation (7) ; dermal (7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Organotin compounds produce neurotoxic and immunotoxic effects. Organotins may directly activate glial cells contributing to neuronal cell degeneration by local release of pro-inflammatory cytokines, tumor necrosis factor-_, and/or interleukins. They may also induce apoptosis by direct action on neuronal cells. Organotin compounds stimulate the neuronal release of and/or decrease of neuronal cell uptake of neurotransmitters in brain tissue, including aspartate, GABA, glutamate, norepinephrine, and serotonin. This may be either a contributing factor to or result of the neuronal cell loss. The immunotoxic effects of organotins are characterized by thymic atrophy caused by the suppression of proliferation of immature thymocytes and apoptosis of mature thymocytes. Organotin compounds are believed to exert these effects by suppressing DNA and protein synthesis, inducing the expression of genes involved in apoptosis (such as nur77), and disrupting the regulation of intracellular calcium levels, giving rise to the uncontrolled production of reactive oxygen species, release of cytochrome c to the cytosol, and the proteolytic and nucleolytic cascade of apoptosis. The suppression of proliferation of immature thymocytes further results in the suppression of T-cell-mediated immune responses. Organotins are also endocrine disruptors and are believed to contribute to obesity by inappropriate receptor activation, leading to adipocyte differentiation. Inorganic tin triggers eryptosis, contributing to tin-induced anemia. (7, 1, 2) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Though tin metal is very poorly absorbed, tin compounds may be absorbed via oral, inhalation, or dermal routes, with organotin compounds being much more readily absorbed than inorganic tin compounds. Tin may enter the bloodstream and bind to hemoglobin, where it is distributed and accumulates mainly in the kidney, liver, lung, and bone. Organotin compounds may undergo dealkylation, hydroxylation, dearylation, and oxidation catalyzed by cytochrome P-450 enzymes in the liver. The alkyl products of dealkylation are conjugated with glutathione and further metabolized to mercapturic acid derivatives. Tin and its metabolites are excreted mainly in the urine and feces. (7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 46 mg/kg (Oral, Rat) (3) LC50: 60.3 mg/m3 over 4 hours (Inhalation, Rat) (4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | This is a man-made compound that is used as a pesticide. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Breathing or swallowing, or skin contact with organotins, can interfere with the way the brain and nervous system work, causing death in severe cases. Organic tin compounds may also damage the immune and reproductive system. (6, 7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Inorganic or organic tin compounds placed on the skin or in the eyes can produce skin and eye irritation. (7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB31790 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 9907219 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 8082871 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C18729 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 30473 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | C007567 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Triphenyltin hydroxide | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | 1440 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D1252.pdf | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity).
- Gene Name:
- PPARG
- Uniprot ID:
- P37231
- Molecular Weight:
- 57619.58 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.02 uM | ATG_PPARg_TRANS | Attagene |
| AC50 | 0.02 uM | ATG_PPARg_TRANS | Attagene |
| AC50 | 0.54 uM | NVS_NR_hPPARg | Novascreen |
References
- Grun F, Blumberg B: Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006 Jun;147(6 Suppl):S50-5. Epub 2006 May 11. [16690801 ]
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1A
- Uniprot ID:
- P07327
- Molecular Weight:
- 39858.37 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1B
- Uniprot ID:
- P00325
- Molecular Weight:
- 39854.21 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1C
- Uniprot ID:
- P00326
- Molecular Weight:
- 39867.27 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH4
- Uniprot ID:
- P08319
- Molecular Weight:
- 40221.335 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH6
- Uniprot ID:
- P28332
- Molecular Weight:
- 39088.335 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Could function in retinol oxidation for the synthesis of retinoic acid, a hormone important for cellular differentiation. Medium-chain (octanol) and aromatic (m-nitrobenzaldehyde) compounds are the best substrates. Ethanol is not a good substrate but at the high ethanol concentrations reached in the digestive tract, it plays a role in the ethanol oxidation and contributes to the first pass ethanol metabolism.
- Gene Name:
- ADH7
- Uniprot ID:
- P40394
- Molecular Weight:
- 41480.985 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Class-III ADH is remarkably ineffective in oxidizing ethanol, but it readily catalyzes the oxidation of long-chain primary alcohols and the oxidation of S-(hydroxymethyl) glutathione.
- Gene Name:
- ADH5
- Uniprot ID:
- P11766
- Molecular Weight:
- 39723.945 Da
References
- Bychkov PV, Shekhovtsova TN, Milaeva ER: Inhibition of horse liver alcohol dehydrogenase by methyltin compounds. Bioinorg Chem Appl. 2005:191-9. doi: 10.1155/BCA.2005.191. [18365099 ]
- General Function:
- Threonine-type endopeptidase activity
- Specific Function:
- The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH. The proteasome has an ATP-dependent proteolytic activity. This unit is responsible of the chymotrypsin-like activity of the proteasome and is one of the principal target of the proteasome inhibitor bortezomib. May catalyze basal processing of intracellular antigens. Plays a role in the protection against oxidative damage through the Nrf2-ARE pathway (By similarity).
- Gene Name:
- PSMB5
- Uniprot ID:
- P28074
- Molecular Weight:
- 28480.01 Da
References
- Shi G, Chen D, Zhai G, Chen MS, Cui QC, Zhou Q, He B, Dou QP, Jiang G: The proteasome is a molecular target of environmental toxic organotins. Environ Health Perspect. 2009 Mar;117(3):379-86. doi: 10.1289/ehp.11865. Epub 2008 Oct 23. [19337512 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. The high affinity ligand for RXRs is 9-cis retinoic acid. RXRA serves as a common heterodimeric partner for a number of nuclear receptors. The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence of ligand, the RXR-RAR heterodimers associate with a multiprotein complex containing transcription corepressors that induce histone acetylation, chromatin condensation and transcriptional suppression. On ligand binding, the corepressors dissociate from the receptors and associate with the coactivators leading to transcriptional activation. The RXRA/PPARA heterodimer is required for PPARA transcriptional activity on fatty acid oxidation genes such as ACOX1 and the P450 system genes.
- Gene Name:
- RXRA
- Uniprot ID:
- P19793
- Molecular Weight:
- 50810.835 Da
References
- Nakanishi T, Nishikawa J, Hiromori Y, Yokoyama H, Koyanagi M, Takasuga S, Ishizaki J, Watanabe M, Isa S, Utoguchi N, Itoh N, Kohno Y, Nishihara T, Tanaka K: Trialkyltin compounds bind retinoid X receptor to alter human placental endocrine functions. Mol Endocrinol. 2005 Oct;19(10):2502-16. Epub 2005 Jun 7. [15941851 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence of ligand, the RXR-RAR heterodimers associate with a multiprotein complex containing transcription corepressors that induce histone acetylation, chromatin condensation and transcriptional suppression. On ligand binding, the corepressors dissociate from the receptors and associate with the coactivators leading to transcriptional activation. RARA plays an essential role in the regulation of retinoic acid-induced germ cell development during spermatogenesis. Has a role in the survival of early spermatocytes at the beginning prophase of meiosis. In Sertoli cells, may promote the survival and development of early meiotic prophase spermatocytes. In concert with RARG, required for skeletal growth, matrix homeostasis and growth plate function (By similarity). Regulates expression of target genes in a ligand-dependent manner by recruiting chromatin complexes containing KMT2E/MLL5. Mediates retinoic acid-induced granulopoiesis.
- Gene Name:
- RARA
- Uniprot ID:
- P10276
- Molecular Weight:
- 50770.805 Da
References
- Grun F, Blumberg B: Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006 Jun;147(6 Suppl):S50-5. Epub 2006 May 11. [16690801 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence or presence of hormone ligand, acts mainly as an activator of gene expression due to weak binding to corepressors. In concert with RARG, required for skeletal growth, matrix homeostasis and growth plate function.
- Gene Name:
- RARB
- Uniprot ID:
- P10826
- Molecular Weight:
- 50488.63 Da
References
- Grun F, Blumberg B: Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006 Jun;147(6 Suppl):S50-5. Epub 2006 May 11. [16690801 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5. In the absence of ligand, acts mainly as an activator of gene expression due to weak binding to corepressors. Required for limb bud development. In concert with RARA or RARB, required for skeletal growth, matrix homeostasis and growth plate function (By similarity).
- Gene Name:
- RARG
- Uniprot ID:
- P13631
- Molecular Weight:
- 50341.405 Da
References
- Grun F, Blumberg B: Environmental obesogens: organotins and endocrine disruption via nuclear receptor signaling. Endocrinology. 2006 Jun;147(6 Suppl):S50-5. Epub 2006 May 11. [16690801 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety. Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. May be required for the propagation of clock information to metabolic pathways regulated by PER2.
- Gene Name:
- PPARA
- Uniprot ID:
- Q07869
- Molecular Weight:
- 52224.595 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.01 uM | ATG_PPRE_CIS | Attagene |
| AC50 | 0.01 uM | ATG_PPRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Receptor for retinoic acid. Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes. The RAR/RXR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5 (By similarity). Specifically binds 9-cis retinoic acid (9C-RA).
- Gene Name:
- RXRB
- Uniprot ID:
- P28702
- Molecular Weight:
- 56921.38 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.01 uM | ATG_RXRb_TRANS | Attagene |
| AC50 | 0.01 uM | ATG_RXRb_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii distal enhancer sequence-specific binding
- Specific Function:
- Transcription activator that binds to antioxidant response (ARE) elements in the promoter regions of target genes. Important for the coordinated up-regulation of genes in response to oxidative stress. May be involved in the transcriptional activation of genes of the beta-globin cluster by mediating enhancer activity of hypersensitive site 2 of the beta-globin locus control region.
- Gene Name:
- NFE2L2
- Uniprot ID:
- Q16236
- Molecular Weight:
- 67825.9 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.02 uM | ATG_NRF2_ARE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Transcriptional regulator which is important for the differentiation and maintenance of meso-diencephalic dopaminergic (mdDA) neurons during development. It is crucial for expression of a set of genes such as SLC6A3, SLC18A2, TH and DRD2 which are essential for development of mdDA neurons (By similarity).
- Gene Name:
- NR4A2
- Uniprot ID:
- P43354
- Molecular Weight:
- 66590.375 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.02 uM | ATG_NURR1_TRANS | Attagene |
| AC50 | 0.02 uM | ATG_NURR1_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- High-capacity urate exporter functioning in both renal and extrarenal urate excretion. Plays a role in porphyrin homeostasis as it is able to mediates the export of protoporhyrin IX (PPIX) both from mitochondria to cytosol and from cytosol to extracellular space, and cellular export of hemin, and heme. Xenobiotic transporter that may play an important role in the exclusion of xenobiotics from the brain. Appears to play a major role in the multidrug resistance phenotype of several cancer cell lines. Implicated in the efflux of numerous drugs and xenobiotics: mitoxantrone, the photosensitizer pheophorbide, camptothecin, methotrexate, azidothymidine (AZT), and the anthracyclines daunorubicin and doxorubicin.
- Gene Name:
- ABCG2
- Uniprot ID:
- Q9UNQ0
- Molecular Weight:
- 72313.47 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.05 uM | CLZD_ABCG2_48 | CellzDirect |
| AC50 | 0.05 uM | CLZD_ABCG2_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- G-protein coupled amine receptor activity
- Specific Function:
- Dopamine receptor whose activity is mediated by G proteins which activate adenylyl cyclase.
- Gene Name:
- DRD1
- Uniprot ID:
- P21728
- Molecular Weight:
- 49292.765 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.15 uM | NVS_GPCR_hDRD1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Voltage-gated calcium channel activity
- Specific Function:
- Receptor for endogenous opioids such as beta-endorphin and endomorphin. Receptor for natural and synthetic opioids including morphine, heroin, DAMGO, fentanyl, etorphine, buprenorphin and methadone. Agonist binding to the receptor induces coupling to an inactive GDP-bound heterotrimeric G-protein complex and subsequent exchange of GDP for GTP in the G-protein alpha subunit leading to dissociation of the G-protein complex with the free GTP-bound G-protein alpha and the G-protein beta-gamma dimer activating downstream cellular effectors. The agonist- and cell type-specific activity is predominantly coupled to pertussis toxin-sensitive G(i) and G(o) G alpha proteins, GNAI1, GNAI2, GNAI3 and GNAO1 isoforms Alpha-1 and Alpha-2, and to a lesser extend to pertussis toxin-insensitive G alpha proteins GNAZ and GNA15. They mediate an array of downstream cellular responses, including inhibition of adenylate cyclase activity and both N-type and L-type calcium channels, activation of inward rectifying potassium channels, mitogen-activated protein kinase (MAPK), phospholipase C (PLC), phosphoinositide/protein kinase (PKC), phosphoinositide 3-kinase (PI3K) and regulation of NF-kappa-B. Also couples to adenylate cyclase stimulatory G alpha proteins. The selective temporal coupling to G-proteins and subsequent signaling can be regulated by RGSZ proteins, such as RGS9, RGS17 and RGS4. Phosphorylation by members of the GPRK subfamily of Ser/Thr protein kinases and association with beta-arrestins is involved in short-term receptor desensitization. Beta-arrestins associate with the GPRK-phosphorylated receptor and uncouple it from the G-protein thus terminating signal transduction. The phosphorylated receptor is internalized through endocytosis via clathrin-coated pits which involves beta-arrestins. The activation of the ERK pathway occurs either in a G-protein-dependent or a beta-arrestin-dependent manner and is regulated by agonist-specific receptor phosphorylation. Acts as a class A G-protein coupled receptor (GPCR) which dissociates from beta-arrestin at or near the plasma membrane and undergoes rapid recycling. Receptor down-regulation pathways are varying with the agonist and occur dependent or independent of G-protein coupling. Endogenous ligands induce rapid desensitization, endocytosis and recycling whereas morphine induces only low desensitization and endocytosis. Heterooligomerization with other GPCRs can modulate agonist binding, signaling and trafficking properties. Involved in neurogenesis. Isoform 12 couples to GNAS and is proposed to be involved in excitatory effects. Isoform 16 and isoform 17 do not bind agonists but may act through oligomerization with binding-competent OPRM1 isoforms and reduce their ligand binding activity.
- Gene Name:
- OPRM1
- Uniprot ID:
- P35372
- Molecular Weight:
- 44778.855 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.18 uM | NVS_GPCR_hOpiate_mu | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins that stimulate adenylate cyclase.
- Gene Name:
- HTR7
- Uniprot ID:
- P34969
- Molecular Weight:
- 53554.43 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.25 uM | NVS_GPCR_h5HT7 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii transcription factor binding
- Specific Function:
- Transcription factor that recognizes and binds to the enhancer heptamer motif 5'-TGA[CG]TCA-3'. Promotes activity of NR5A1 when phosphorylated by HIPK3 leading to increased steroidogenic gene expression upon cAMP signaling pathway stimulation.
- Gene Name:
- JUN
- Uniprot ID:
- P05412
- Molecular Weight:
- 35675.32 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.29 uM | CLM_StressKinase_1hr | Cellumen |
| AC50 | 0.28 uM | CLM_StressKinase_24hr | Cellumen |
| AC50 | 0.28 uM | CLM_StressKinase_24hr | Cellumen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- TP53
- Uniprot ID:
- P04637
- Molecular Weight:
- 43652.79 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.02 uM | CLM_p53Act_1hr | Cellumen |
| AC50 | 0.39 uM | CLM_p53Act_72hr | Cellumen |
| AC50 | 0.39 uM | CLM_p53Act_72hr | Cellumen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Histone binding
- Specific Function:
- Variant histone H2A which replaces conventional H2A in a subset of nucleosomes. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling. Required for checkpoint-mediated arrest of cell cycle progression in response to low doses of ionizing radiation and for efficient repair of DNA double strand breaks (DSBs) specifically when modified by C-terminal phosphorylation.
- Gene Name:
- H2AFX
- Uniprot ID:
- P16104
- Molecular Weight:
- 15144.45 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.42 uM | CLM_OxidativeStress_1hr | Cellumen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein homodimerization activity
- Specific Function:
- Alpha-2 adrenergic receptors mediate the catecholamine-induced inhibition of adenylate cyclase through the action of G proteins.
- Gene Name:
- ADRA2C
- Uniprot ID:
- P18825
- Molecular Weight:
- 49521.585 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.43 uM | NVS_GPCR_hAdra2C | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Opioid receptor activity
- Specific Function:
- G-protein coupled receptor that functions as receptor for endogenous enkephalins and for a subset of other opioids. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling leads to the inhibition of adenylate cyclase activity. Inhibits neurotransmitter release by reducing calcium ion currents and increasing potassium ion conductance. Plays a role in the perception of pain and in opiate-mediated analgesia. Plays a role in developing analgesic tolerance to morphine.
- Gene Name:
- OPRD1
- Uniprot ID:
- P41143
- Molecular Weight:
- 40368.235 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.46 uM | NVS_GPCR_hOpiate_D1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Sh3 domain binding
- Specific Function:
- Dopamine receptor responsible for neuronal signaling in the mesolimbic system of the brain, an area of the brain that regulates emotion and complex behavior. Its activity is mediated by G proteins which inhibit adenylyl cyclase. Modulates the circadian rhythm of contrast sensitivity by regulating the rhythmic expression of NPAS2 in the retinal ganglion cells (By similarity).
- Gene Name:
- DRD4
- Uniprot ID:
- P21917
- Molecular Weight:
- 48359.86 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.73 uM | NVS_GPCR_hDRD4.4 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
- Specific Function:
- Gtp binding
- Gene Name:
- TUBA1A
- Uniprot ID:
- Q71U36
- Molecular Weight:
- 50135.25 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.82 uM | CLM_MicrotubuleCSK_1hr | Cellumen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein tyrosine/serine/threonine phosphatase activity
- Specific Function:
- Shows activity both for tyrosine-protein phosphate and serine-protein phosphate, but displays a strong preference toward phosphotyrosines. Specifically dephosphorylates and inactivates ERK1 and ERK2.
- Gene Name:
- DUSP3
- Uniprot ID:
- P51452
- Molecular Weight:
- 20478.1 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.83 uM | NVS_ENZ_hDUSP3 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds and is activated by variety of endogenous and xenobiotic compounds. Transcription factor that activates the transcription of multiple genes involved in the metabolism and secretion of potentially harmful xenobiotics, drugs and endogenous compounds. Activated by the antibiotic rifampicin and various plant metabolites, such as hyperforin, guggulipid, colupulone, and isoflavones. Response to specific ligands is species-specific. Activated by naturally occurring steroids, such as pregnenolone and progesterone. Binds to a response element in the promoters of the CYP3A4 and ABCB1/MDR1 genes.
- Gene Name:
- NR1I2
- Uniprot ID:
- O75469
- Molecular Weight:
- 49761.245 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.86 uM | NVS_NR_hPXR | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor signaling protein activity
- Specific Function:
- Beta-adrenergic receptors mediate the catecholamine-induced activation of adenylate cyclase through the action of G proteins. This receptor binds epinephrine and norepinephrine with approximately equal affinity. Mediates Ras activation through G(s)-alpha- and cAMP-mediated signaling.
- Gene Name:
- ADRB1
- Uniprot ID:
- P08588
- Molecular Weight:
- 51322.1 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.00 uM | NVS_GPCR_hAdrb1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Norepinephrine:sodium symporter activity
- Specific Function:
- Amine transporter. Terminates the action of noradrenaline by its high affinity sodium-dependent reuptake into presynaptic terminals.
- Gene Name:
- SLC6A2
- Uniprot ID:
- P23975
- Molecular Weight:
- 69331.42 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.10 uM | NVS_TR_hNET | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Potassium channel regulator activity
- Specific Function:
- Dopamine receptor whose activity is mediated by G proteins which inhibit adenylyl cyclase.
- Gene Name:
- DRD2
- Uniprot ID:
- P14416
- Molecular Weight:
- 50618.91 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.30 uM | NVS_GPCR_hDRD2s | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Sulfotransferase activity
- Specific Function:
- Sulfotransferase that utilizes 3'-phospho-5'-adenylyl sulfate (PAPS) as sulfonate donor to catalyze the sulfonation of steroids and bile acids in the liver and adrenal glands.
- Gene Name:
- SULT2A1
- Uniprot ID:
- Q06520
- Molecular Weight:
- 33779.57 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.33 uM | CLZD_SULT2A1_6 | CellzDirect |
| AC50 | 4.61 uM | CLZD_SULT2A1_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins.
- Gene Name:
- HTR5A
- Uniprot ID:
- P47898
- Molecular Weight:
- 40254.69 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.80 uM | NVS_GPCR_h5HT5A | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 3.44 uM | CLZD_CYP2B6_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.00 uM | CLZD_CYP3A4_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Responsible for the metabolism of a number of therapeutic agents such as the anticonvulsant drug S-mephenytoin, omeprazole, proguanil, certain barbiturates, diazepam, propranolol, citalopram and imipramine.
- Gene Name:
- CYP2C19
- Uniprot ID:
- P33261
- Molecular Weight:
- 55930.545 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.10 uM | NVS_ADME_hCYP2C19 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transporter activity
- Specific Function:
- Involved in the ATP-dependent secretion of bile salts into the canaliculus of hepatocytes.
- Gene Name:
- ABCB11
- Uniprot ID:
- O95342
- Molecular Weight:
- 146405.83 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.21 uM | CLZD_ABCB11_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Most active in catalyzing 2-hydroxylation. Caffeine is metabolized primarily by cytochrome CYP1A2 in the liver through an initial N3-demethylation. Also acts in the metabolism of aflatoxin B1 and acetaminophen. Participates in the bioactivation of carcinogenic aromatic and heterocyclic amines. Catalizes the N-hydroxylation of heterocyclic amines and the O-deethylation of phenacetin.
- Gene Name:
- CYP1A2
- Uniprot ID:
- P05177
- Molecular Weight:
- 58293.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.03 uM | CLZD_CYP1A2_6 | CellzDirect |
| AC50 | 5.59 uM | CLZD_CYP1A2_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Glutathione transferase activity
- Specific Function:
- Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles.
- Gene Name:
- GSTA2
- Uniprot ID:
- P09210
- Molecular Weight:
- 25663.675 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.65 uM | CLZD_GSTA2_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Hydroxymethylglutaryl-coa synthase activity
- Specific Function:
- This enzyme condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, which is the substrate for HMG-CoA reductase.
- Gene Name:
- HMGCS2
- Uniprot ID:
- P54868
- Molecular Weight:
- 56634.915 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 6.24 uM | CLZD_HMGCS2_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serotonin receptor activity
- Specific Function:
- This is one of the several different receptors for 5-hydroxytryptamine (serotonin), a biogenic hormone that functions as a neurotransmitter, a hormone, and a mitogen. The activity of this receptor is mediated by G proteins that stimulate adenylate cyclase. It has a high affinity for tricyclic psychotropic drugs (By similarity). Controls pyramidal neurons migration during corticogenesis, through the regulation of CDK5 activity (By similarity). Is an activator of TOR signaling (PubMed:23027611).
- Gene Name:
- HTR6
- Uniprot ID:
- P50406
- Molecular Weight:
- 46953.625 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.60 uM | NVS_GPCR_h5HT6 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. In the epoxidation of arachidonic acid it generates only 14,15- and 11,12-cis-epoxyeicosatrienoic acids. It is the principal enzyme responsible for the metabolism the anti-cancer drug paclitaxel (taxol).
- Gene Name:
- CYP2C8
- Uniprot ID:
- P10632
- Molecular Weight:
- 55824.275 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.60 uM | NVS_ADME_hCYP2C8 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d 24-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP1A1
- Uniprot ID:
- P04798
- Molecular Weight:
- 58164.815 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.19 uM | CLZD_CYP1A1_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]