alpha-Amanitin (T3D2450)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-07-03 17:43:41 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:25:31 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D2450 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | alpha-Amanitin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | alpha-Amanitin or α-amanitin is a cyclic peptide of eight amino acids. It is possibly the most deadly of all the amatoxins, toxins found in several members of the Amanita genus of mushrooms, one being the Death cap (Amanita phalloides) as well as the Destroying angel, a complex of similar species, principally A. virosa and A. bisporigera. It is also found in the mushrooms Galerina marginata and Conocybe filaris. (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
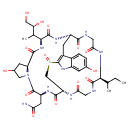
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C39H54N10O14S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 918.970 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 918.354 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 23109-05-9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 2-[(1R,4S,10S,13S,16S,27R,34S)-34-(butan-2-yl)-13-[(3R)-3,4-dihydroxybutan-2-yl]-8,22-dihydroxy-2,5,11,14,27,30,33,36,39-nonaoxo-27λ⁴-thia-3,6,12,15,25,29,32,35,38-nonaazapentacyclo[14.12.11.0⁶,¹⁰.0¹⁸,²⁶.0¹⁹,²⁴]nonatriaconta-18(26),19,21,23-tetraen-4-yl]acetamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | 2-[(1R,4S,10S,13S,16S,27R,34S)-13-[(3R)-3,4-dihydroxybutan-2-yl]-8,22-dihydroxy-2,5,11,14,27,30,33,36,39-nonaoxo-34-(sec-butyl)-27λ⁴-thia-3,6,12,15,25,29,32,35,38-nonaazapentacyclo[14.12.11.0⁶,¹⁰.0¹⁸,²⁶.0¹⁹,²⁴]nonatriaconta-18(26),19,21,23-tetraen-4-yl]acetamide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CCC(C)[C@@H]1NC(=O)CNC(=O)[C@@H]2CC3=C(NC4=CC(O)=CC=C34)[S@](=O)C[C@H](NC(=O)CNC1=O)C(=O)N[C@@H](CC(N)=O)C(=O)N1CC(O)C[C@H]1C(=O)N[C@@H](C(C)[C@@H](O)CO)C(=O)N2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C39H54N10O14S/c1-4-16(2)31-36(60)42-11-29(55)43-25-15-64(63)38-21(20-6-5-18(51)7-22(20)46-38)9-23(33(57)41-12-30(56)47-31)44-37(61)32(17(3)27(53)14-50)48-35(59)26-8-19(52)13-49(26)39(62)24(10-28(40)54)45-34(25)58/h5-7,16-17,19,23-27,31-32,46,50-53H,4,8-15H2,1-3H3,(H2,40,54)(H,41,57)(H,42,60)(H,43,55)(H,44,61)(H,45,58)(H,47,56)(H,48,59)/t16?,17?,19?,23-,24-,25-,26-,27-,31-,32-,64+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=CIORWBWIBBPXCG-NUCBJAHASA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of inorganic compounds known as alkaline earth metal bromides. These are inorganic compounds in which the largest halogen atom is Bromine, and the heaviest metal atom a lanthanide. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Inorganic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Mixed metal/non-metal compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Alkaline earth metal salts | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alkaline earth metal bromides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alkaline earth metal bromides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Liquid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Clear solution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Oral, dermal, inhalation, and parenteral (contaminated drugs). (2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Amanitin has an unusually strong and specific attraction to the enzyme RNA polymerase II. Upon ingestion, it binds to the RNA polymerase II enzyme, preventing mRNA synthesis and effectively causing cytolysis of hepatocytes (liver cells). (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Free toxin may be removed by opsonization via the reticuloendothelial system (primarily the liver and kidneys) or it may be degraded through cellular internalization via the lysosomes. Lysosomes are membrane-enclosed organelles that contain an array of digestive enzymes, including several proteases. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 0.1 mg/kg (Oral, Mouse) (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | It is found in several members of the Amanita genus of mushrooms, one being the Death cap (Amanita phalloides) as well as the Destroying angel, a complex of similar species, principally A. virosa and A. bisporigera. It is also found in the mushrooms Galerina marginata and Conocybe filaris. (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Around 15% of those poisoned will die within 10 days, progressing through a comatose stage to renal failure, liver failure, hepatic coma, respiratory failure and death. Those who recover are at risk of permanent liver damage. Diagnosis is difficult, and is established by observation of the clinical symptoms as well as the presence of α-amanitin in the urine. Urine screening is generally most useful within 48 hours of ingestion. (1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Few effects are reported within 10 hours; it is not unusual for significant effects to take as long as 24 hours after ingestion to appear, with this delay in symptoms making α-amanitin poisoning even more difficult to diagnose and all the more dangerous. By then, it is far past the time in which stomach pumping would yield an efficient result. Diarrhea and cramps are the first symptoms, but those pass, giving a false sign of remission. Typically, on the 4th to 5th day, the toxin starts to have severe effects on the liver and kidneys, leading to total system failure in both. Death usually takes place around a week from ingestion. (3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Treatment is mainly supportive (gastric lavage, activated carbon, fluid resuscitation) but includes various drugs to counter the amatoxins, including intravenous penicillin and cephalosporin derivatives, and, in cases of greater ingestion, can extend to an orthotopic liver transplant. The most reliable method to treat amanitin poisoning is through having the stomach pumped immediately after ingestion; however, the onset of symptoms is generally too late for this to be an option. (1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 2100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C08438 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | A8W7M4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 37415 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | alpha-Amanitin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Alpha-amanitin | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D2450.pdf | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Largest and catalytic component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Forms the polymerase active center together with the second largest subunit. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB1 is part of the core element with the central large cleft, the clamp element that moves to open and close the cleft and the jaws that are thought to grab the incoming DNA template. At the start of transcription, a single-stranded DNA template strand of the promoter is positioned within the central active site cleft of Pol II. A bridging helix emanates from RPB1 and crosses the cleft near the catalytic site and is thought to promote translocation of Pol II by acting as a ratchet that moves the RNA-DNA hybrid through the active site by switching from straight to bent conformations at each step of nucleotide addition. During transcription elongation, Pol II moves on the template as the transcript elongates. Elongation is influenced by the phosphorylation status of the C-terminal domain (CTD) of Pol II largest subunit (RPB1), which serves as a platform for assembly of factors that regulate transcription initiation, elongation, termination and mRNA processing. Acts as an RNA-dependent RNA polymerase when associated with small delta antigen of Hepatitis delta virus, acting both as a replicate and transcriptase for the viral RNA circular genome.
- Gene Name:
- POLR2A
- Uniprot ID:
- P24928
- Molecular Weight:
- 217174.235 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Lrr domain binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB11 is part of the core element with the central large cleft (By similarity).
- Gene Name:
- POLR2J
- Uniprot ID:
- P52435
- Molecular Weight:
- 13293.19 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB11 is part of the core element with the central large cleft (By similarity).
- Gene Name:
- POLR2J2
- Uniprot ID:
- Q9GZM3
- Molecular Weight:
- 13088.14 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB11 is part of the core element with the central large cleft (By similarity).
- Gene Name:
- POLR2J3
- Uniprot ID:
- Q9H1A7
- Molecular Weight:
- 13092.11 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Ribonucleoside binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Second largest component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Proposed to contribute to the polymerase catalytic activity and forms the polymerase active center together with the largest subunit. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB2 is part of the core element with the central large cleft, the clamp element that moves to open and close the cleft and the jaws that are thought to grab the incoming DNA template (By similarity).
- Gene Name:
- POLR2B
- Uniprot ID:
- P30876
- Molecular Weight:
- 133895.435 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB3 is part of the core element with the central large cleft and the clamp element that moves to open and close the cleft (By similarity).
- Gene Name:
- POLR2C
- Uniprot ID:
- P19387
- Molecular Weight:
- 31440.86 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Translation initiation factor binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB4 is part of a subcomplex with RPB7 that binds to a pocket formed by RPB1, RPB2 and RPB6 at the base of the clamp element. The RBP4-RPB7 subcomplex seems to lock the clamp via RPB7 in the closed conformation thus preventing double-stranded DNA to enter the active site cleft. The RPB4-RPB7 subcomplex binds single-stranded DNA and RNA (By similarity).
- Gene Name:
- POLR2D
- Uniprot ID:
- O15514
- Molecular Weight:
- 16311.105 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Translation initiation factor binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB7 is part of a subcomplex with RPB4 that binds to a pocket formed by RPB1, RPB2 and RPB6 at the base of the clamp element. The RBP4-RPB7 subcomplex seems to lock the clamp via RPB7 in the closed conformation thus preventing double-stranded DNA to enter the active site cleft. The RPB4-RPB7 subcomplex binds single-stranded DNA and RNA (By similarity). Binds RNA.
- Gene Name:
- POLR2G
- Uniprot ID:
- P62487
- Molecular Weight:
- 19294.195 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Zinc ion binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. Pol II is the central component of the basal RNA polymerase II transcription machinery. It is composed of mobile elements that move relative to each other. RPB9 is part of the upper jaw surrounding the central large cleft and thought to grab the incoming DNA template (By similarity).
- Gene Name:
- POLR2I
- Uniprot ID:
- P36954
- Molecular Weight:
- 14523.1 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common component of RNA polymerases I, II and III which synthesize ribosomal RNA precursors, mRNA precursors and many functional non-coding RNAs, and small RNAs, such as 5S rRNA and tRNAs, respectively. Pol II is the central component of the basal RNA polymerase II transcription machinery. Pols are composed of mobile elements that move relative to each other. In Pol II, POLR2E/RPB5 is part of the lower jaw surrounding the central large cleft and thought to grab the incoming DNA template. Seems to be the major component in this process (By similarity).
- Gene Name:
- POLR2E
- Uniprot ID:
- P19388
- Molecular Weight:
- 24551.075 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerases catalyze the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common component of RNA polymerases I, II, and III which synthesize ribosomal RNA precursors, mRNA precursors and many functional non-coding RNAs, and small RNAs, such as 5S rRNA and tRNAs, respectively. Pol II is the central component of the basal RNA polymerase II transcription machinery. Pols are composed of mobile elements that move relative to each other. In Pol II, POLR2F/RPB6 is part of the clamp element and together with parts of RPB1 and RPB2 forms a pocket to which the RPB4-RPB7 subcomplex binds (By similarity).
- Gene Name:
- POLR2F
- Uniprot ID:
- P61218
- Molecular Weight:
- 14477.92 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Dna-directed rna polymerase activity
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common component of RNA polymerases I, II and III which synthesize ribosomal RNA precursors, mRNA precursors and many functional non-coding RNAs, and small RNAs, such as 5S rRNA and tRNAs, respectively.
- Gene Name:
- POLR2H
- Uniprot ID:
- P52434
- Molecular Weight:
- 17143.115 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Zinc ion binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common component of RNA polymerases I, II and III which synthesize ribosomal RNA precursors, mRNA precursors and many functional non-coding RNAs, and a small RNAs, such as 5S rRNA and tRNAs, respectively.
- Gene Name:
- POLR2K
- Uniprot ID:
- P53803
- Molecular Weight:
- 7004.145 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]
- General Function:
- Zinc ion binding
- Specific Function:
- DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. Common component of RNA polymerases I, II and III which synthesize ribosomal RNA precursors, mRNA precursors and many functional non-coding RNAs, and a small RNAs, such as 5S rRNA and tRNAs, respectively. Pol II is the central component of the basal RNA polymerase II transcription machinery. Pols are composed of mobile elements that move relative to each other. In Pol II, POLR2L/RBP10 is part of the core element with the central large cleft (By similarity).
- Gene Name:
- POLR2L
- Uniprot ID:
- P62875
- Molecular Weight:
- 7645.02 Da
References
- Wikipedia. alpha-Amanitin. Last Updated 25 May 2009. [Link]