| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-05 03:20:59 UTC |
|---|
| Update Date | 2014-12-24 20:25:42 UTC |
|---|
| Accession Number | T3D2568 |
|---|
| Identification |
|---|
| Common Name | Ceftriaxone |
|---|
| Class | Small Molecule |
|---|
| Description | Ceftriaxone is only found in individuals that have used or taken this drug. It is a broad-spectrum cephalosporin antibiotic with a very long half-life and high penetrability to meninges, eyes and inner ears. [PubChem]Ceftriaxone works by inhibiting the mucopeptide synthesis in the bacterial cell wall. The beta-lactam moiety of Ceftriaxone binds to carboxypeptidases, endopeptidases, and transpeptidases in the bacterial cytoplasmic membrane. These enzymes are involved in cell-wall synthesis and cell division. By binding to these enzymes, Ceftriaxone results in the formation of of defective cell walls and cell death. |
|---|
| Compound Type | - Amide
- Amine
- Anti-Bacterial Agent
- Cephalosporin
- Drug
- Ester
- Ether
- Metabolite
- Organic Compound
- Synthetic Compound
|
|---|
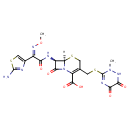
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (6R,7R)-7-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-3-{[(2-methyl-5,6-dioxo-1,2,5,6-tetrahydro-1,2,4-triazin-3-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid | | Acantex | | Biotrakson | | Cefatriaxone | | Ceftriaxona | | Ceftriaxonum | | Ceftriazone | | Rocephin | | Rocephine |
|
|---|
| Chemical Formula | C18H18N8O7S3 |
|---|
| Average Molecular Mass | 554.580 g/mol |
|---|
| Monoisotopic Mass | 554.046 g/mol |
|---|
| CAS Registry Number | 73384-59-5 |
|---|
| IUPAC Name | (6R,7R)-7-[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetamido]-3-{[(2-methyl-5,6-dioxo-1,2,5,6-tetrahydro-1,2,4-triazin-3-yl)sulfanyl]methyl}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid |
|---|
| Traditional Name | ceftriaxone |
|---|
| SMILES | [H][C@]12SCC(CSC3=NC(=O)C(=O)NN3C)=C(N1C(=O)[C@@]2([H])N=C(O)C(=N/OC)\C1=CSC(=N)N1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H18N8O7S3/c1-25-18(22-12(28)13(29)23-25)36-4-6-3-34-15-9(14(30)26(15)10(6)16(31)32)21-11(27)8(24-33-2)7-5-35-17(19)20-7/h5,9,15H,3-4H2,1-2H3,(H2,19,20)(H,21,27)(H,23,29)(H,31,32)/b24-8-/t9-,15-/m1/s1 |
|---|
| InChI Key | InChIKey=VAAUVRVFOQPIGI-SPQHTLEESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as aminoglycosides. These are molecules or a portion of a molecule composed of amino-modified sugars. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Aminoglycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aminoglycoside core
- Macrolide
- Glycosyl compound
- O-glycosyl compound
- Oxane
- Monosaccharide
- Tertiary alcohol
- 1,2-aminoalcohol
- Amino acid or derivatives
- Carboxylic acid ester
- Ketone
- Cyclic ketone
- Tertiary aliphatic amine
- Tertiary amine
- Secondary alcohol
- Lactone
- Acetal
- Carboxylic acid derivative
- Dialkyl ether
- Ether
- Oxacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Alcohol
- Amine
- Organopnictogen compound
- Organonitrogen compound
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | Solid (MSDS, A308). |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | >155°C | | Boiling Point | Not Available | | Solubility | 1.05e-01 g/L | | LogP | -1.7 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-0924040000-11c1c4ec5dc051494298 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fr-9774123000-c094d2c5681b0477a4aa | 2017-10-06 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Ceftriaxone,1TMS,#1" TMS) - 70eV, Positive | Not Available | 2021-10-14 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | 2021-10-15 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0apl-3069470000-ea9533b06f6d727b78d9 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05mk-6139010000-862d2dcbf27819f7fdaa | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9043100000-fbf004668a64dff18475 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4r-5691220000-22e1520a7886282ef024 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4l-4940000000-3a6002666fe076cf5ad8 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-160408d1a7b4d7ae1995 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0013090000-bb4e8b82e0fb04e55aad | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0avi-0466190000-080d962933aef409d2d1 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056u-1944420000-18694b9d401854bf9e80 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0pb9-0910080000-bcb5ef0d6c356695310e | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-803717215c9d04868fa4 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9400000000-75fd909b2cc72f5d0010 | 2021-10-11 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Inhalation; dermal or skin contact; ingestion; intravenous (MSDS, A308). |
|---|
| Mechanism of Toxicity | Ceftriaxone works by inhibiting the mucopeptide synthesis in the bacterial cell wall. The beta-lactam moiety of Ceftriaxone binds to carboxypeptidases, endopeptidases, and transpeptidases in the bacterial cytoplasmic membrane. These enzymes are involved in cell-wall synthesis and cell division. By binding to these enzymes, Ceftriaxone results in the formation of of defective cell walls and cell death. |
|---|
| Metabolism | Ceftriaxone is eliminated unchanged in the urine by glomerular filtration (60%) and bile (40%) (2).
Route of Elimination: Thirty-three percent to 67% of a ceftriaxone dose was excreted in the urine as unchanged drug and the remainder was secreted in the bile and ultimately found in the feces as microbiologically inactive compounds.
Half Life: 5.8-8.7 hours |
|---|
| Toxicity Values | LD50: >10 000 mg/kg (Oral, Rat) |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | For the treatment of the infections (respiratory, skin, soft tissue, UTI, ENT) caused by S. pneumoniae, H. influenzae, staphylococci, S. pyogenes (group A beta-hemolytic streptococci), E. coli, P. mirabilis, Klebsiella sp, coagulase-negative staph (1). |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Some observed adverse reactions ( < 0.1%) include abdominal pain, agranulocytosis, allergic pneumonitis, anaphylaxis, basophilia, biliary lithiasis, bronchospasm, colitis, dyspepsia, epistaxis, flatulence, gallbladder sludge, glycosuria, hematuria, jaundice, leukocytosis, lymphocytosis, monocytosis, nephrolithiasis, palpitations, a decrease in the prothrombin time, renal precipitations, seizures, and serum sickness (RxList, A308). |
|---|
| Symptoms | Abdominal pain. |
|---|
| Treatment | In the case of overdosage, drug concentration would not be reduced by hemodialysis or peritoneal dialysis. There is no specific antidote. Treatment of overdosage should be symptomatic (RxList, A308). |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB01212 |
|---|
| HMDB ID | HMDB15343 |
|---|
| PubChem Compound ID | 5479530 |
|---|
| ChEMBL ID | CHEMBL161 |
|---|
| ChemSpider ID | 4514885 |
|---|
| KEGG ID | C06683 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 29007 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Ceftriaxone |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Ceftriaxone |
|---|
| References |
|---|
| Synthesis Reference | Monguzzi Riccardo, Menaspace Silvano, Anzaghi Piergiorgio, “Process for the preparation of ceftriaxone.” U.S. Patent US5026843, issued November, 1984. |
|---|
| MSDS | Link |
|---|
| General References | - Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M: DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901-6. Epub 2007 Nov 29. [18048412 ]
- Miolo G, Caffieri S, Levorato L, Imbesi M, Giusti P, Uz T, Manev R, Manev H: Photoisomerization of fluvoxamine generates an isomer that has reduced activity on the 5-hydroxytryptamine transporter and does not affect cell proliferation. Eur J Pharmacol. 2002 Aug 30;450(3):223-9. [12208313 ]
- Brittain HG and Florey K. Profiles of Drug Substances, Excipients and Related Methodology. Volume 30. Toronto ON: Elsevier.
- The Merck Index (2009). Ceftriaxone. [Link]
- Drugs.com [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|