| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-21 20:27:51 UTC |
|---|
| Update Date | 2014-12-24 20:25:53 UTC |
|---|
| Accession Number | T3D2918 |
|---|
| Identification |
|---|
| Common Name | Diflunisal |
|---|
| Class | Small Molecule |
|---|
| Description | Diflunisal, a salicylate derivative, is a nonsteroidal anti-inflammatory agent (NSAIA) with pharmacologic actions similar to other prototypical NSAIAs. Diflunisal possesses anti-inflammatory, analgesic and antipyretic activity. Though its mechanism of action has not been clearly established, most of its actions appear to be associated with inhibition of prostaglandin synthesis via the arachidonic acid pathway. Diflunisal is used to relieve pain accompanied with inflammation and in the symptomatic treatment of rheumatoid arthritis and osteoarthritis. |
|---|
| Compound Type | - Anti-Inflammatory Agent, Non-Steroidal
- Cyclooxygenase Inhibitor
- Drug
- Ester
- Metabolite
- Organic Compound
- Organofluoride
- Synthetic Compound
|
|---|
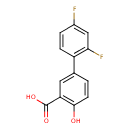
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 2',4'-Difluoro-4-hydroxy-3-biphenylcarboxylic acid | | 2-(Hydroxy)-5-(2,4-difluorophenyl)benzoic acid | | 5-(2,4-Difluorophenyl)salicylic acid | | Anton | | Diflunisalum | | Dolobid |
|
|---|
| Chemical Formula | C13H8F2O3 |
|---|
| Average Molecular Mass | 250.198 g/mol |
|---|
| Monoisotopic Mass | 250.044 g/mol |
|---|
| CAS Registry Number | 22494-42-4 |
|---|
| IUPAC Name | 5-(2,4-difluorophenyl)-2-hydroxybenzoic acid |
|---|
| Traditional Name | diflunisal |
|---|
| SMILES | OC(=O)C1=C(O)C=CC(=C1)C1=C(F)C=C(F)C=C1 |
|---|
| InChI Identifier | InChI=1S/C13H8F2O3/c14-8-2-3-9(11(15)6-8)7-1-4-12(16)10(5-7)13(17)18/h1-6,16H,(H,17,18) |
|---|
| InChI Key | InChIKey=HUPFGZXOMWLGNK-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as biphenyls and derivatives. These are organic compounds containing to benzene rings linked together by a C-C bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Biphenyls and derivatives |
|---|
| Direct Parent | Biphenyls and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Biphenyl
- Hydroxybenzoic acid
- Salicylic acid or derivatives
- Salicylic acid
- Benzoic acid or derivatives
- Benzoic acid
- Benzoyl
- 1-hydroxy-2-unsubstituted benzenoid
- Fluorobenzene
- Halobenzene
- Phenol
- Aryl fluoride
- Aryl halide
- Vinylogous acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Organohalogen compound
- Organofluoride
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | | Name | SMPDB Link | KEGG Link |
|---|
| Diflunisal Pathway | Not Available | Not Available |
|
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 210-211°C | | Boiling Point | Not Available | | Solubility | Practically insoluble (14.5 mg/L) at neutral or acidic pH. | | LogP | 4.44 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pc0-1190000000-37ea5d58644c01ef5b1e | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-05fr-5019000000-223530358c78521d8313 | 2017-10-06 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-2690000000-64056c9dc4e2f14d3c59 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4j-5590000000-462ea99995ea9238bf94 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4i-5490000000-ec23e03578a95364b748 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0a4i-9550000000-2fa065aea811fa7b6c5a | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000t-9000000000-01d2c7c75ab3756b3df7 | 2017-09-14 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0090000000-e1009fb8e100faed2065 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f89-0090000000-e9bcdf3ba0212ee28f45 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0fc0-1970000000-baed3c5d64825d6b04b2 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-052b-0090000000-6427bf95a0116aa621f1 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0090000000-83af00a8dd308410951d | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0690000000-a756057c6442130830ef | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-0090000000-645b3ae2575f11227813 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0090000000-78fd25facb031cdb643f | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-0940000000-9920437171fca6936f28 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0090000000-3e0ed3bddcaf7a500b10 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052b-0090000000-4ac100e6866826f4e00a | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0kdi-1930000000-6c51e381123d99125d70 | 2021-10-11 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0fai-1690000000-d231ff5a9ac7d9769d2d | 2014-09-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Rapidly and completely absorbed following oral administration, with a bioavailability of 80-90%. Peak plasma concentrations are achieved 2 - 3 hours following oral administration. |
|---|
| Mechanism of Toxicity | The precise mechanism of the analgesic and anti-inflammatory actions of diflunisal is not known. Diflunisal is a prostaglandin synthetase inhibitor. In animals, prostaglandins sensitize afferent nerves and potentiate the action of bradykinin in inducing pain. Since prostaglandins are known to be among the mediators of pain and inflammation, the mode of action of diflunisal may be due to a decrease of prostaglandins in peripheral tissues. |
|---|
| Metabolism | Hepatic, primarily via glucuronide conjugation (90% of administered dose).
Route of Elimination: The drug is excreted in the urine as two soluble glucuronide conjugates accounting for about 90% of the administered dose. Little or no diflunisal is excreted in the feces.
Half Life: 8 to 12 hours |
|---|
| Toxicity Values | LD50: 392 mg/kg (Oral, Rat) (1)
LD50: 439 mg/kg (Oral, Mouse) (1)
LD50: 603 mg/kg (Oral, Rabbit) (1) |
|---|
| Lethal Dose | The lowest dose without the presence of other medicines which caused death was 15 grams. |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Used to relieve pain, tenderness, swelling and stiffness caused by osteoarthritis (arthritis caused by a breakdown of the lining of the joints) and rheumatoid arthritis (arthritis caused by swelling of the lining of the joints). [L1869] |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Symptoms of overdose include coma, tachycardia, stupor, and vomiting. |
|---|
| Treatment | In the event of overdosage, the stomach should be emptied by inducing vomiting or by gastric lavage, and the patient carefully observed and given symptomatic and supportive treatment. Because of the high degree of protein binding, hemodialysis may not be effective. (3) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00861 |
|---|
| HMDB ID | HMDB14999 |
|---|
| PubChem Compound ID | 3059 |
|---|
| ChEMBL ID | CHEMBL898 |
|---|
| ChemSpider ID | 2951 |
|---|
| KEGG ID | C01691 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 39669 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Diflunisal |
|---|
| PDB ID | 1FL |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Diflunisal |
|---|
| References |
|---|
| Synthesis Reference | Ruyle, W.V., Jarett, L.H. and Matzuk, A.R. ; U S . Patent 3,714,226; January 30, 1973; assigned to Merck & Co., Inc. |
|---|
| MSDS | Link |
|---|
| General References | - Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M: DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901-6. Epub 2007 Nov 29. [18048412 ]
- Medline Plus. Diflunisal [Link]
- RxList: The Internet Drug Index (2009). [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | | Gene | Gene Symbol | Gene ID | Interaction | Chromosome | Details |
|---|
|
|---|
| Down-Regulated Genes | Not Available |
|---|