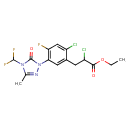| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2013-04-25 07:56:50 UTC |
|---|
| Update Date | 2014-12-24 20:26:32 UTC |
|---|
| Accession Number | T3D3805 |
|---|
| Identification |
|---|
| Common Name | Carfentrazone-ethyl |
|---|
| Class | Small Molecule |
|---|
| Description | Carfentrazone-ethyl is a contact herbicide used to control broadleaf and sedge weeds in cereals. The mode of action of carfentrazone-ethyl is the disruption of membranes by inhibiting the action of protoporphyrinogen oxidase, causing cell death. Carfentrazone is non-selective, and can be used for complete vegetation control, as well as as a desiccant and a defoliant in some crops. |
|---|
| Compound Type | - Ester
- Ether
- Herbicide
- Organic Compound
- Organochloride
- Organofluoride
- Pesticide
- Synthetic Compound
|
|---|
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | Affinity | | Annex | | Aurora | | Ethyl 2-chloro-3-(2-chloro-5-[4-(difluoromethyl)-3-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-4-fluorophenyl)propanoate | | Spotlight |
|
|---|
| Chemical Formula | C15H14Cl2F3N3O3 |
|---|
| Average Molecular Mass | 412.191 g/mol |
|---|
| Monoisotopic Mass | 411.036 g/mol |
|---|
| CAS Registry Number | 128639-02-1 |
|---|
| IUPAC Name | ethyl 2-chloro-3-{2-chloro-5-[4-(difluoromethyl)-3-methyl-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-4-fluorophenyl}propanoate |
|---|
| Traditional Name | carfentrazone-ethyl |
|---|
| SMILES | CCOC(=O)C(Cl)CC1=C(Cl)C=C(F)C(=C1)N1N=C(C)N(C(F)F)C1=O |
|---|
| InChI Identifier | InChI=1S/C15H14Cl2F3N3O3/c1-3-26-13(24)10(17)4-8-5-12(11(18)6-9(8)16)23-15(25)22(14(19)20)7(2)21-23/h5-6,10,14H,3-4H2,1-2H3 |
|---|
| InChI Key | InChIKey=MLKCGVHIFJBRCD-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenyl-1,2,4-triazoles. These are organic compounds containing a 1,2,4-triazole substituted by a phenyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Azoles |
|---|
| Sub Class | Triazoles |
|---|
| Direct Parent | Phenyl-1,2,4-triazoles |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenyl-1,2,4-triazole
- Chlorobenzene
- Fatty acid ester
- Fluorobenzene
- Halobenzene
- Aryl chloride
- Aryl fluoride
- Aryl halide
- Monocyclic benzene moiety
- Benzenoid
- Fatty acyl
- Heteroaromatic compound
- Alpha-halocarboxylic acid or derivatives
- Alpha-halocarboxylic acid derivative
- Carboxylic acid ester
- Azacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxide
- Carbonyl group
- Organohalogen compound
- Alkyl halide
- Alkyl fluoride
- Alkyl chloride
- Organooxygen compound
- Hydrocarbon derivative
- Organochloride
- Organofluoride
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004j-9234000000-7fb4e7e380bf8e0a2d43 | 2021-09-24 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 45V, Positive | splash10-0fba-0195000000-30d06b61935e7ffbdcc2 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 60V, Positive | splash10-00bc-0490000000-e386d7b27e56da4eb555 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-00kb-0009000000-afc535c3cbf8dea68646 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 15V, Positive | splash10-03yi-0009600000-a552ec509d50ae8e0d98 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 75V, Positive | splash10-00ou-0940000000-fafe9c790cea3eb58eb7 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 90V, Positive | splash10-00mo-0910000000-e9a8378e4d00552e1205 | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1209500000-dda3116936ad8755bc19 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9113000000-b558b0dc60e2060a28a9 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-7920000000-c0d0b986c4c4c16748b4 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-08c964d8471330a35c21 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xr-9316000000-ec86f68cc29f8bd196e8 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9000000000-8ea6e2e15eef36d82df1 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0003900000-ede933b8dd68d899a8db | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-0009000000-e6b4624f66e3500b78fe | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000f-3597000000-9239c3ed1faa2e8609bd | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0022900000-b36e79db94173bb2f621 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-0090000000-bc64fa22a027cc77270e | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0v4j-6967000000-dd6d1596788c7cb98141 | 2021-10-12 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-03ec-1429100000-3b153ce061eafe11bb29 | 2014-10-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | LD50 (rat, oral) >5000 mg/kg
LD50 (rat, dermal) >4000 mg/kg
LC50 (rat, inhalation) >5.09 mg/L |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | This is a man-made compound that is used as a pesticide. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Carfentrazone-ethyl is not considered to cause significant toxicity in helathy humans. But in patient with very large acute exposures or long-term chronic exposures, carfentrazone-ethyl may cause variegate porphyria or transient porphyria cutanea tarda-like syndrome. (1) |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Remove contaminated clothing and wash skin with soap and water. For eye exposure, copious irrigation with water or saline is used. The treatment is usually symptomatic and supportive. |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 86222 |
|---|
| ChEMBL ID | CHEMBL2145069 |
|---|
| ChemSpider ID | 77773 |
|---|
| KEGG ID | C11094 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D3805.pdf |
|---|
| General References | - Toxnet - Carfentrazone-ethyl [Link]
- EPA - Carfentrazone-ethyl [Link]
- ENVIRONMENTAL PROTECTION AGENCY
Environmental Protection Agency - Carfentrazone-ethyl [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|