| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2013-04-25 07:56:50 UTC |
|---|
| Update Date | 2014-12-24 20:26:32 UTC |
|---|
| Accession Number | T3D3806 |
|---|
| Identification |
|---|
| Common Name | Chloridazon |
|---|
| Class | Small Molecule |
|---|
| Description | Chloridazon is a selective herbicide belonging to the group of pyridazone - derivatives , which was put on the market by BASF in the 1960s and used mainly in beet cultivation to control annual broad-leafed weeds. It acts by inhibiting photosynthesis and the Hill reaction and is rapidly absorbed through the roots of plants with tranlocation acropetally to all plant parts. |
|---|
| Compound Type | - Amide
- Amine
- Ester
- Herbicide
- Organic Compound
- Organochloride
- Pesticide
- Synthetic Compound
|
|---|
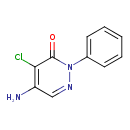
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 1-Phenyl-4-amino-5-chloro-6-pyridazone | | 1-Phenyl-4-amino-5-chloropyridaz-6-one | | 1-Phenyl-4-amino-5-chlorpyridaz-6-one | | 5-Amino-4-chloro-2,3-dihydro-3-oxo-2-phenylpyridazine | | 5-amino-4-chloro-2-phenyl-2,3-dihydropyridazin-3-one | | 5-Amino-4-chloro-2-phenyl-3(2H)-pyridazinone | | 5-Amino-4-chloro-2-phenyl-3(2H)-pyridazone | | 5-Amino-4-chloro-2-phenyl-3-pyridazinone | | 5-Amino-4-chloro-2-phenyl-pyridazin-3-one | | 5-amino-4-chloro-2-phenylpyridazin-3(2H)-one | | 5-Amino-4-chloro-2-phenylpyridazinone | | Alicep | | Betoxon | | Burex | | Choridazone | | Clorizol | | PCA | | Phenazon | | Phenazone | | Pyramin | | Pyramine | | Pyrazone | | Suzon | | Tripart gladiator |
|
|---|
| Chemical Formula | C10H8ClN3O |
|---|
| Average Molecular Mass | 221.643 g/mol |
|---|
| Monoisotopic Mass | 221.036 g/mol |
|---|
| CAS Registry Number | 1698-60-8 |
|---|
| IUPAC Name | 5-amino-4-chloro-2-phenyl-2,3-dihydropyridazin-3-one |
|---|
| Traditional Name | chloridazon |
|---|
| SMILES | NC1=C(Cl)C(=O)N(N=C1)C1=CC=CC=C1 |
|---|
| InChI Identifier | InChI=1S/C10H8ClN3O/c11-9-8(12)6-13-14(10(9)15)7-4-2-1-3-5-7/h1-6H,12H2 |
|---|
| InChI Key | InChIKey=WYKYKTKDBLFHCY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyridazinones. Pyridazinones are compounds containing a pyridazine ring which bears a ketone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyridazines and derivatives |
|---|
| Direct Parent | Pyridazinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aminopyridazine
- Pyridazinone
- Aryl chloride
- Aryl halide
- Monocyclic benzene moiety
- Benzenoid
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Azacycle
- Amine
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organochloride
- Organohalogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0190000000-499ab271b2220d999ccc | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0190000000-e94c9fa1186c77da4154 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-51ce14b4d625e2244bf9 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0290000000-cb9ca1a52809502bde50 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0uk9-1960000000-8054f0e2cf7b35a123a6 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9800000000-4b5b9c0ddf6f6ce12650 | 2016-08-03 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-00fr-9150000000-169ea799def095dc3980 | 2014-10-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | This is a man-made compound that is used as a pesticide. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 15546 |
|---|
| ChEMBL ID | CHEMBL1525705 |
|---|
| ChemSpider ID | 14790 |
|---|
| KEGG ID | C18570 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D3806.pdf |
|---|
| General References | Not Available |
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|