Monomethyl phthalate (T3D3886)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2013-04-25 07:56:53 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:33 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D3886 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Monomethyl phthalate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Monomethyl phthalate is a metabolite, or breakdown product, of dimethyl phthalate, used in insect repellant, plastic, and as a rocket propellant. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
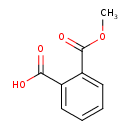
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C9H8O4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 180.157 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 180.042 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 4376-18-5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 2-(methoxycarbonyl)benzoic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | methyl hydrogen phthalate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | COC(=O)C1=CC=CC=C1C(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C9H8O4/c1-13-9(12)7-5-3-2-4-6(7)8(10)11/h2-5H,1H3,(H,10,11) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=FNJSWIPFHMKRAT-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as benzoic acid esters. These are ester derivatives of benzoic acid. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Benzenoids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzene and substituted derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Benzoic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Benzoic acid esters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic homomonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Monomethyl phthalate is a metabolite, or breakdown product, of dimethyl phthalate, used in insect repellant, plastic, and as a rocket propellant. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB02130 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 20392 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL243609 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 19207 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 491486 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | D3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Medonos, Vladimir; Pasek, Josef; Ruzicka, Vlastimil. Preparation of methyl phthalate by addition of dimethyl ether to phthalic anhydride. Chemicky Prumysl (1957), 7 281-5. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Zinc ion binding
- Specific Function:
- Ligand-activated transcription factor. Key regulator of lipid metabolism. Activated by the endogenous ligand 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Activated by oleylethanolamide, a naturally occurring lipid that regulates satiety. Receptor for peroxisome proliferators such as hypolipidemic drugs and fatty acids. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the ACOX1 and P450 genes. Transactivation activity requires heterodimerization with RXRA and is antagonized by NR2C2. May be required for the propagation of clock information to metabolic pathways regulated by PER2.
- Gene Name:
- PPARA
- Uniprot ID:
- Q07869
- Molecular Weight:
- 52224.595 Da
References
- Sarath Josh MK, Pradeep S, Vijayalekshmi Amma KS, Balachandran S, Abdul Jaleel UC, Doble M, Spener F, Benjamin S: Phthalates efficiently bind to human peroxisome proliferator activated receptor and retinoid X receptor alpha, beta, gamma subtypes: an in silico approach. J Appl Toxicol. 2014 Jul;34(7):754-65. doi: 10.1002/jat.2902. Epub 2013 Jul 11. [23843199 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Ligand-activated transcription factor. Receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Has a preference for poly-unsaturated fatty acids, such as gamma-linoleic acid and eicosapentanoic acid. Once activated by a ligand, the receptor binds to promoter elements of target genes. Regulates the peroxisomal beta-oxidation pathway of fatty acids. Functions as transcription activator for the acyl-CoA oxidase gene. Decreases expression of NPC1L1 once activated by a ligand.
- Gene Name:
- PPARD
- Uniprot ID:
- Q03181
- Molecular Weight:
- 49902.99 Da
References
- Sarath Josh MK, Pradeep S, Vijayalekshmi Amma KS, Balachandran S, Abdul Jaleel UC, Doble M, Spener F, Benjamin S: Phthalates efficiently bind to human peroxisome proliferator activated receptor and retinoid X receptor alpha, beta, gamma subtypes: an in silico approach. J Appl Toxicol. 2014 Jul;34(7):754-65. doi: 10.1002/jat.2902. Epub 2013 Jul 11. [23843199 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds peroxisome proliferators such as hypolipidemic drugs and fatty acids. Once activated by a ligand, the nuclear receptor binds to DNA specific PPAR response elements (PPRE) and modulates the transcription of its target genes, such as acyl-CoA oxidase. It therefore controls the peroxisomal beta-oxidation pathway of fatty acids. Key regulator of adipocyte differentiation and glucose homeostasis. ARF6 acts as a key regulator of the tissue-specific adipocyte P2 (aP2) enhancer. Acts as a critical regulator of gut homeostasis by suppressing NF-kappa-B-mediated proinflammatory responses. Plays a role in the regulation of cardiovascular circadian rhythms by regulating the transcription of ARNTL/BMAL1 in the blood vessels (By similarity).
- Gene Name:
- PPARG
- Uniprot ID:
- P37231
- Molecular Weight:
- 57619.58 Da
References
- Sarath Josh MK, Pradeep S, Vijayalekshmi Amma KS, Balachandran S, Abdul Jaleel UC, Doble M, Spener F, Benjamin S: Phthalates efficiently bind to human peroxisome proliferator activated receptor and retinoid X receptor alpha, beta, gamma subtypes: an in silico approach. J Appl Toxicol. 2014 Jul;34(7):754-65. doi: 10.1002/jat.2902. Epub 2013 Jul 11. [23843199 ]
- General Function:
- Hydroxymethylglutaryl-coa synthase activity
- Specific Function:
- This enzyme condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, which is the substrate for HMG-CoA reductase.
- Gene Name:
- HMGCS2
- Uniprot ID:
- P54868
- Molecular Weight:
- 56634.915 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.05 uM | CLZD_HMGCS2_6 | CellzDirect |
| AC50 | 0.05 uM | CLZD_HMGCS2_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. This enzyme contributes to the wide pharmacokinetics variability of the metabolism of drugs such as S-warfarin, diclofenac, phenytoin, tolbutamide and losartan.
- Gene Name:
- CYP2C9
- Uniprot ID:
- P11712
- Molecular Weight:
- 55627.365 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.35 uM | CLZD_CYP2C9_6 | CellzDirect |
| AC50 | 0.35 uM | CLZD_CYP2C9_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Cleaves collagens of types I, II, and III at one site in the helical domain. Also cleaves collagens of types VII and X. In case of HIV infection, interacts and cleaves the secreted viral Tat protein, leading to a decrease in neuronal Tat's mediated neurotoxicity.
- Gene Name:
- MMP1
- Uniprot ID:
- P03956
- Molecular Weight:
- 54006.61 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_MMP1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serine-type endopeptidase inhibitor activity
- Specific Function:
- Serine protease inhibitor. This inhibitor acts as 'bait' for tissue plasminogen activator, urokinase, protein C and matriptase-3/TMPRSS7. Its rapid interaction with PLAT may function as a major control point in the regulation of fibrinolysis.
- Gene Name:
- SERPINE1
- Uniprot ID:
- P05121
- Molecular Weight:
- 45059.695 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_PAI1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]