| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-08-28 21:19:25 UTC |
|---|
| Update Date | 2014-12-24 20:26:34 UTC |
|---|
| Accession Number | T3D3955 |
|---|
| Identification |
|---|
| Common Name | Tetracycline |
|---|
| Class | Small Molecule |
|---|
| Description | Tetracycline is a broad spectrum polyketide antibiotic produced by the Streptomyces genus of Actinobacteria. It exerts a bacteriostatic effect on bacteria by binding reversible to the bacterial 30S ribosomal subunit and blocking incoming aminoacyl tRNA from binding to the ribosome acceptor site. It also binds to some extent to the bacterial 50S ribosomal subunit and may alter the cytoplasmic membrane causing intracellular components to leak from bacterial cells. |
|---|
| Compound Type | - Anti-Bacterial Agent
- Antiprotozoal Agent
- Drug
- Metabolite
- Organic Compound
- Protein Synthesis Inhibitor
- Synthetic Compound
- Tetracycline
|
|---|
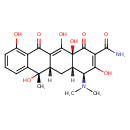
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (4S,4AS,5as,12as)-4-(dimethylamino)-1,4,4a,5,5a,6,11,12a-octahydro-3,6,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-2-naphthacenecarboxamide | | Abramycin | | Achromycin | | Anhydrotetracycline | | Deschlorobiomycin | | Liquamycin | | Tetracyclin | | TETRACYCLINE | | Tetracycline HCl | | Tetracyclinum | | Tetrazyklin | | Tsiklomitsin |
|
|---|
| Chemical Formula | C22H24N2O8 |
|---|
| Average Molecular Mass | 444.435 g/mol |
|---|
| Monoisotopic Mass | 444.153 g/mol |
|---|
| CAS Registry Number | 60-54-8 |
|---|
| IUPAC Name | (4S,4aS,5aS,6S,12aS)-4-(dimethylamino)-3,6,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide |
|---|
| Traditional Name | tetracycline |
|---|
| SMILES | [H][C@]12C[C@@]3([H])[C@]([H])(N(C)C)C(O)=C(C(O)=N)C(=O)[C@@]3(O)C(O)=C1C(=O)C1=C(C=CC=C1O)[C@@]2(C)O |
|---|
| InChI Identifier | InChI=1S/C22H24N2O8/c1-21(31)8-5-4-6-11(25)12(8)16(26)13-9(21)7-10-15(24(2)3)17(27)14(20(23)30)19(29)22(10,32)18(13)28/h4-6,9-10,15,25,27-28,31-32H,7H2,1-3H3,(H2,23,30)/t9-,10-,15-,21+,22-/m0/s1 |
|---|
| InChI Key | InChIKey=OFVLGDICTFRJMM-WESIUVDSSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tetracyclines. These are polyketides having an octahydrotetracene-2-carboxamide skeleton, substituted with many hydroxy and other groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Phenylpropanoids and polyketides |
|---|
| Class | Tetracyclines |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Tetracyclines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tetracycline
- Naphthacene
- Tetracene
- Anthracene carboxylic acid or derivatives
- Tetralin
- Aryl ketone
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Cyclohexenone
- Aralkylamine
- Benzenoid
- Tertiary alcohol
- Vinylogous acid
- Tertiary aliphatic amine
- Amino acid or derivatives
- Tertiary amine
- Carboxamide group
- Primary carboxylic acid amide
- Ketone
- Polyol
- Carboxylic acid derivative
- Enol
- Amine
- Organic oxygen compound
- Organopnictogen compound
- Organonitrogen compound
- Organooxygen compound
- Alcohol
- Carbonyl group
- Organic nitrogen compound
- Organic oxide
- Hydrocarbon derivative
- Aromatic homopolycyclic compound
|
|---|
| Molecular Framework | Aromatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | |
|---|
| Pathways | | Name | SMPDB Link | KEGG Link |
|---|
| Tetracycline Action Pathway | SMP00294 | Not Available |
|
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 172.5 dec°C | | Boiling Point | Not Available | | Solubility | 231 mg/L (at 25°C) | | LogP | -1.3 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0bvi-5259500000-bd5eacef407c7cbcfc2a | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0002-5023749000-5781179b3d4ec0f18e56 | 2017-10-06 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_1) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_2) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_3) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_8) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_9) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_10) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_5_1) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_5_2) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_5_3) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_5_8) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_1) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_5) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_6) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_7) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Tetracycline,2TMS,#2" TMS) - 70eV, Positive | Not Available | 2021-10-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-01sc-1900000000-7b792268aeb0a63c32c5 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-01p9-1900000000-da0b69e206bb0369723c | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-000i-3900000000-299e4496b1f8e2f213cc | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-01ta-0000900000-e5ca6594fe72eb2bcd1f | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0ik9-0402900000-9ef4a0d08180ce9b2a1c | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-0ik9-0402900000-a84ce5e2767545f4dd7e | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-01sc-1900000000-7b792268aeb0a63c32c5 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 15V, Positive | splash10-01ta-0000900000-908fcaa876a610a2ecbe | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 45V, Negative | splash10-01p9-1900000000-da0b69e206bb0369723c | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004j-0000900000-b0b8239482dc0ab660e1 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01t9-0001900000-11d8380f89252d777068 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0btc-2268900000-9b20bc13399f86c0ff4a | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0002900000-bbde63c16f1d6207b45a | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fal-0118900000-1d65f8ee8fa0af312c20 | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00r6-9166000000-84828062c581777db668 | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0000900000-3355d21a81297e70a696 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0000900000-aec34c9f29d5643c9dd7 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-015i-9333100000-24fa0a1d06d44e57511d | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002f-0004900000-ce47e08c76771852979b | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05ec-0049700000-8b1cbde084e892a26b15 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001u-1069300000-553a5cff4aef98461f76 | 2021-10-11 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Bioavailability is less than 40% when administered via intramuscular injection, 100% intravenously, and 60-80% orally (fasting adults). Food and/or milk reduce GI absorption of oral preparations of tetracycline by 50% or more. |
|---|
| Mechanism of Toxicity | Tetracyclines target the 28S small subunit of the mitochondrial ribosome thereby deactivation mitochondrial protein synthesis. As a result tetracyclines are cytotoxic to the most metabolically active cells or tissues including the heart, liver, thymus and bone-marrow. (2). The likely target of most tetracyclines is the 12S rRNA molecule in the mitochondrial ribosome, which is analogous to the 16S rRNA in bacterial ribosomes. |
|---|
| Metabolism | Not metabolized
Route of Elimination: They are concentrated by the liver in the bile and excreted in the urine and feces at high concentrations in a biologically active form.
Half Life: 6-12 hours |
|---|
| Toxicity Values | LD50=808mg/kg (orally in mice) |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Used to treat bacterial infections such as Rocky Mountain spotted fever, typhus fever, tick fevers, Q fever, rickettsialpox and Brill-Zinsser disease. May be used to treat infections caused by Chlamydiae spp., B. burgdorferi (Lyme disease), and upper respiratory infections caused by typical (S. pneumoniae, H. influenzae, and M. catarrhalis) and atypical organisms (C. pneumoniae, M. pneumoniae, L. pneumophila). May also be used to treat acne. Tetracycline may be an alternative drug for people who are allergic to penicillin. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Side effects from normal doses of tetracyclines are relatively minimal, but of particular note is phototoxicity. Tetracylclines increase the risk of sunburn under exposure to light from the sun or other sources. Tetracyclines may also cause stomach or bowel upsets, and, on rare occasions, allergic reactions. Very rarely, severe headache and vision problems may be signs of dangerous secondary intracranial hypertension, also known as pseudotumor cerebri. Tetracyclines are teratogens and cause tooth discolouration and poor tooth mineralization in the fetus as they develop in infancy. Symptoms of tetracycline overdose include anorexia, nausea, diarrhea, glossitis, dysphagia, enterocolitis and inflammatory lesions, skin reactions such as maculopapular and erythematous rashes, exfoliative dermatitis, photosensitivity, hypersensitivity reactions such as urticaria, angioneurotic oedema, anaphylaxis, anaphyl-actoid purpura, pericarditis, and exacerbation of systemic lupus erythematosus, benign intracranial hypertension in adults disappearing on discontinuation of the medicine, haematologic abnormalities such as haemolytic anaemia, thrombocytopenia, neutropenia, and eosinophilia. |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Drug therapy is discontinued immediately; exchange transfusion may be required to remove the drug. Sometimes, phenobarbital (UGT induction) is used. |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00759 |
|---|
| HMDB ID | HMDB14897 |
|---|
| PubChem Compound ID | 5280962 |
|---|
| ChEMBL ID | CHEMBL1440 |
|---|
| ChemSpider ID | 4510286 |
|---|
| KEGG ID | C06570 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 27902 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Tetracycline |
|---|
| References |
|---|
| Synthesis Reference | Thomas F. McNamara, Nungavaram S. Ramamurthy, Lorne M. Golub, “Non-antibacterial tetracycline compositions possessing anti-collagenolytic properties and methods of preparing and using same.” U.S. Patent US4704383, issued May, 1963. |
|---|
| MSDS | Link |
|---|
| General References | - Griffin MO, Fricovsky E, Ceballos G, Villarreal F: Tetracyclines: a pleitropic family of compounds with promising therapeutic properties. Review of the literature. Am J Physiol Cell Physiol. 2010 Sep;299(3):C539-48. doi: 10.1152/ajpcell.00047.2010. Epub 2010 Jun 30. [20592239 ]
- McKee EE, Ferguson M, Bentley AT, Marks TA: Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother. 2006 Jun;50(6):2042-9. [16723564 ]
- Wikipedia. Tetracycline. Last updated on 29 July 2014. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|