Adenosine triphosphate (T3D4276)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2014-08-29 06:12:10 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:45 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D4276 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Adenosine triphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Adenosine triphosphate (ATP) is a nucleotide consisting of a purine base (adenine) attached to the first carbon atom of ribose (a pentose sugar). Three phosphate groups are esterified at the fifth carbon atom of the ribose. ATP is incorporated into nucleic acids by polymerases in the processes of DNA replication and transcription. ATP contributes to cellular energy charge and participates in overall energy balance, maintaining cellular homeostasis. ATP can act as an extracellular signaling molecule via interactions with specific purinergic receptors to mediate a wide variety of processes as diverse as neurotransmission, inflammation, apoptosis, and bone remodelling. Extracellular ATP and its metabolite adenosine have also been shown to exert a variety of effects on nearly every cell type in human skin, and ATP seems to play a direct role in triggering skin inflammatory, regenerative, and fibrotic responses to mechanical injury, an indirect role in melanocyte proliferation and apoptosis, and a complex role in Langerhans cell-directed adaptive immunity. During exercise, intracellular homeostasis depends on the matching of adenosine triphosphate (ATP) supply and ATP demand. Metabolites play a useful role in communicating the extent of ATP demand to the metabolic supply pathways. Effects as different as proliferation or differentiation, chemotaxis, release of cytokines or lysosomal constituents, and generation of reactive oxygen or nitrogen species are elicited upon stimulation of blood cells with extracellular ATP. The increased concentration of adenosine triphosphate (ATP) in erythrocytes from patients with chronic renal failure (CRF) has been observed in many studies but the mechanism leading to these abnormalities still is controversial. (1, 2, 3, 4, 5). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
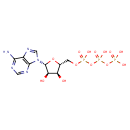
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C10H16N5O13P3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 507.181 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 506.996 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 56-65-5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | ({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | adenosine triphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@]1(COP(O)(=O)OP(O)(=O)OP(O)(O)=O)O[C@@]([H])(N2C=NC3=C(N)N=CN=C23)[C@]([H])(O)[C@]1([H])O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C10H16N5O13P3/c11-8-5-9(13-2-12-8)15(3-14-5)10-7(17)6(16)4(26-10)1-25-30(21,22)28-31(23,24)27-29(18,19)20/h2-4,6-7,10,16-17H,1H2,(H,21,22)(H,23,24)(H2,11,12,13)(H2,18,19,20)/t4-,6-,7-,10-/m1/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=ZKHQWZAMYRWXGA-KQYNXXCUSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as purine ribonucleoside triphosphates. These are purine ribobucleotides with a triphosphate group linked to the ribose moiety. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside triphosphates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Endogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | ATP is able to store and transport chemical energy within cells. ATP also plays an important role in the synthesis of nucleic acids. ATP can be produced by various cellular processes, most typically in mitochondria by oxidative phosphorylation under the catalytic influence of ATP synthase. The total quantity of ATP in the human body is about 0.1 mole. The energy used by human cells requires the hydrolysis of 200 to 300 moles of ATP daily. This means that each ATP molecule is recycled 2000 to 3000 times during a single day. ATP cannot be stored, hence its consumption must closely follow its synthesis. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Metabolism of organophosphates occurs principally by oxidation, by hydrolysis via esterases and by reaction with glutathione. Demethylation and glucuronidation may also occur. Oxidation of organophosphorus pesticides may result in moderately toxic products. In general, phosphorothioates are not directly toxic but require oxidative metabolism to the proximal toxin. The glutathione transferase reactions produce products that are, in most cases, of low toxicity. Paraoxonase (PON1) is a key enzyme in the metabolism of organophosphates. PON1 can inactivate some organophosphates through hydrolysis. PON1 hydrolyzes the active metabolites in several organophosphates insecticides as well as, nerve agents such as soman, sarin, and VX. The presence of PON1 polymorphisms causes there to be different enzyme levels and catalytic efficiency of this esterase, which in turn suggests that different individuals may be more susceptible to the toxic effect of organophosphate exposure. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Oral LD50 in rats is > 2 g/kg. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | For nutritional supplementation, also for treating dietary shortage or imbalance | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB00171 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB00538 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 5957 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL14249 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 5742 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C00002 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 15422 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | ATP | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | ATP | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Adenosine_triphosphate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | George M. Whitesides, Patricia E. Garrett, Merrell G. Siegel, “Method for preparing adenosine triphosphate.” U.S. Patent US4164444, issued April, 1975. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Toxin transporter activity
- Specific Function:
- Transporter involved in macrophage lipid homeostasis. Is an active component of the macrophage lipid export complex. Could also be involved in intracellular lipid transport processes. The role in cellular lipid homeostasis may not be limited to macrophages.
- Gene Name:
- ABCG1
- Uniprot ID:
- P45844
- Molecular Weight:
- 75591.275 Da
References
- Lage H: MDR1/P-glycoprotein (ABCB1) as target for RNA interference-mediated reversal of multidrug resistance. Curr Drug Targets. 2006 Jul;7(7):813-21. [16842213 ]
- Materna V, Stege A, Surowiak P, Priebsch A, Lage H: RNA interference-triggered reversal of ABCC2-dependent cisplatin resistance in human cancer cells. Biochem Biophys Res Commun. 2006 Sep 15;348(1):153-7. Epub 2006 Jul 14. [16876126 ]
- Thomas AC, Cullup T, Norgett EE, Hill T, Barton S, Dale BA, Sprecher E, Sheridan E, Taylor AE, Wilroy RS, DeLozier C, Burrows N, Goodyear H, Fleckman P, Stephens KG, Mehta L, Watson RM, Graham R, Wolf R, Slavotinek A, Martin M, Bourn D, Mein CA, O'Toole EA, Kelsell DP: ABCA12 is the major harlequin ichthyosis gene. J Invest Dermatol. 2006 Nov;126(11):2408-13. Epub 2006 Aug 10. [16902423 ]
- Girardin F: Membrane transporter proteins: a challenge for CNS drug development. Dialogues Clin Neurosci. 2006;8(3):311-21. [17117613 ]
- Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC: An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007 Jan 19;315(5810):373-7. Epub 2006 Dec 7. [17158291 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- Energy-dependent efflux pump responsible for decreased drug accumulation in multidrug-resistant cells.
- Gene Name:
- ABCB1
- Uniprot ID:
- P08183
- Molecular Weight:
- 141477.255 Da
References
- Lage H: MDR1/P-glycoprotein (ABCB1) as target for RNA interference-mediated reversal of multidrug resistance. Curr Drug Targets. 2006 Jul;7(7):813-21. [16842213 ]
- Gardner ER, Burger H, van Schaik RH, van Oosterom AT, de Bruijn EA, Guetens G, Prenen H, de Jong FA, Baker SD, Bates SE, Figg WD, Verweij J, Sparreboom A, Nooter K: Association of enzyme and transporter genotypes with the pharmacokinetics of imatinib. Clin Pharmacol Ther. 2006 Aug;80(2):192-201. [16890580 ]
- Blume H, Donath F, Warnke A, Schug BS: Pharmacokinetic drug interaction profiles of proton pump inhibitors. Drug Saf. 2006;29(9):769-84. [16944963 ]
- Gervasini G, Carrillo JA, Garcia M, San Jose C, Cabanillas A, Benitez J: Adenosine triphosphate-binding cassette B1 (ABCB1) (multidrug resistance 1) G2677T/A gene polymorphism is associated with high risk of lung cancer. Cancer. 2006 Dec 15;107(12):2850-7. [17120199 ]
- Fukui N, Suzuki Y, Sawamura K, Sugai T, Watanabe J, Inoue Y, Someya T: Dose-dependent effects of the 3435 C>T genotype of ABCB1 gene on the steady-state plasma concentration of fluvoxamine in psychiatric patients. Ther Drug Monit. 2007 Apr;29(2):185-9. [17417072 ]
- General Function:
- Sulfonylurea receptor activity
- Specific Function:
- Subunit of the beta-cell ATP-sensitive potassium channel (KATP). Regulator of ATP-sensitive K(+) channels and insulin release.
- Gene Name:
- ABCC8
- Uniprot ID:
- Q09428
- Molecular Weight:
- 176990.36 Da
References
- Nakamura A, Kawahito S, Kawano T, Nazari H, Takahashi A, Kitahata H, Nakaya Y, Oshita S: Differential effects of etomidate and midazolam on vascular adenosine triphosphate-sensitive potassium channels: isometric tension and patch clamp studies. Anesthesiology. 2007 Mar;106(3):515-22. [17325510 ]
- Bienengraeber M, Warltier DC, Bosnjak ZJ, Stadnicka A: Mechanism of cardiac sarcolemmal adenosine triphosphate-sensitive potassium channel activation by isoflurane in a heterologous expression system. Anesthesiology. 2006 Sep;105(3):534-40. [16931986 ]
- Bryan J, Munoz A, Zhang X, Dufer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L: ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflugers Arch. 2007 Feb;453(5):703-18. Epub 2006 Aug 8. [16897043 ]
- Tanaka K, Kawano T, Nakamura A, Nazari H, Kawahito S, Oshita S, Takahashi A, Nakaya Y: Isoflurane activates sarcolemmal adenosine triphosphate-sensitive potassium channels in vascular smooth muscle cells: a role for protein kinase A. Anesthesiology. 2007 May;106(5):984-91. [17457130 ]
- General Function:
- Atp binding
- Specific Function:
- Activates acetate so that it can be used for lipid synthesis or for energy generation.
- Gene Name:
- ACSS2
- Uniprot ID:
- Q9NR19
- Molecular Weight:
- 78579.11 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC: Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002 Dec 20;298(5602):2390-2. [12493915 ]
- Schuler AJ, Jenkins D: Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, Part II: Anaerobic adenosine triphosphate utilization and acetate uptake rates. Water Environ Res. 2003 Nov-Dec;75(6):499-511. [14704009 ]
- General Function:
- Syntaxin binding
- Specific Function:
- cAMP-dependent and sulfonylurea-sensitive anion transporter. Key gatekeeper influencing intracellular cholesterol transport.
- Gene Name:
- ABCA1
- Uniprot ID:
- O95477
- Molecular Weight:
- 254299.89 Da
References
- Porchay I, Pean F, Bellili N, Royer B, Cogneau J, Chesnier MC, Caradec A, Tichet J, Balkau B, Marre M, Fumeron F: ABCA1 single nucleotide polymorphisms on high-density lipoprotein-cholesterol and overweight: the D.E.S.I.R. study. Obesity (Silver Spring). 2006 Nov;14(11):1874-9. [17135600 ]
- Badeau R, Jauhiainen M, Metso J, Nikander E, Tikkanen MJ, Ylikorkala O, Mikkola TS: Effect of isolated isoflavone supplementation on ABCA1-dependent cholesterol efflux potential in postmenopausal women. Menopause. 2007 Mar-Apr;14(2):293-9. [17224860 ]
- Zarubica A, Trompier D, Chimini G: ABCA1, from pathology to membrane function. Pflugers Arch. 2007 Feb;453(5):569-79. Epub 2006 Jul 21. [16858612 ]
- General Function:
- Transporter activity
- Specific Function:
- Subunit of ATP-sensitive potassium channels (KATP). Can form cardiac and smooth muscle-type KATP channels with KCNJ11. KCNJ11 forms the channel pore while ABCC9 is required for activation and regulation.
- Gene Name:
- ABCC9
- Uniprot ID:
- O60706
- Molecular Weight:
- 174221.7 Da
References
- Zhao JL, Yang YJ, You SJ, Jing ZC, Wu YJ, Cheng JL, Gao RL: Pretreatment with fosinopril or valsartan reduces myocardial no-reflow after acute myocardial infarction and reperfusion. Coron Artery Dis. 2006 Aug;17(5):463-9. [16845255 ]
- Yang YJ, Zhao JL, You SJ, Wu YJ, Jing ZC, Gao RL, Chen ZJ: Post-infarction treatment with simvastatin reduces myocardial no-reflow by opening of the KATP channel. Eur J Heart Fail. 2007 Jan;9(1):30-6. Epub 2006 Jul 7. [16829188 ]
- Bryan J, Munoz A, Zhang X, Dufer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L: ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflugers Arch. 2007 Feb;453(5):703-18. Epub 2006 Aug 8. [16897043 ]
- General Function:
- Receptor binding
- Specific Function:
- Non-receptor tyrosine-protein kinase that plays an ABL1-overlapping role in key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion and receptor endocytosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like MYH10 (involved in movement); CTTN (involved in signaling); or TUBA1 and TUBB (microtubule subunits). Binds directly F-actin and regulates actin cytoskeletal structure through its F-actin-bundling activity. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as CRK, CRKL, DOK1 or ARHGAP35. Adhesion-dependent phosphorylation of ARHGAP35 promotes its association with RASA1, resulting in recruitment of ARHGAP35 to the cell periphery where it inhibits RHO. Phosphorylates multiple receptor tyrosine kinases like PDGFRB and other substrates which are involved in endocytosis regulation such as RIN1. In brain, may regulate neurotransmission by phosphorylating proteins at the synapse. ABL2 acts also as a regulator of multiple pathological signaling cascades during infection. Pathogens can highjack ABL2 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1.
- Gene Name:
- ABL2
- Uniprot ID:
- P42684
- Molecular Weight:
- 128341.935 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- General Function:
- Nucleotide binding
- Specific Function:
- Oligomeric Apaf-1 mediates the cytochrome c-dependent autocatalytic activation of pro-caspase-9 (Apaf-3), leading to the activation of caspase-3 and apoptosis. This activation requires ATP. Isoform 6 is less effective in inducing apoptosis.
- Gene Name:
- APAF1
- Uniprot ID:
- O14727
- Molecular Weight:
- 141838.815 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Chinnaiyan AM: The apoptosome: heart and soul of the cell death machine. Neoplasia. 1999 Apr;1(1):5-15. [10935465 ]
- General Function:
- Organic anion transmembrane transporter activity
- Specific Function:
- Mediates hepatobiliary excretion of numerous organic anions. May function as a cellular cisplatin transporter.
- Gene Name:
- ABCC2
- Uniprot ID:
- Q92887
- Molecular Weight:
- 174205.64 Da
References
- Materna V, Stege A, Surowiak P, Priebsch A, Lage H: RNA interference-triggered reversal of ABCC2-dependent cisplatin resistance in human cancer cells. Biochem Biophys Res Commun. 2006 Sep 15;348(1):153-7. Epub 2006 Jul 14. [16876126 ]
- Rau T, Erney B, Gores R, Eschenhagen T, Beck J, Langer T: High-dose methotrexate in pediatric acute lymphoblastic leukemia: impact of ABCC2 polymorphisms on plasma concentrations. Clin Pharmacol Ther. 2006 Nov;80(5):468-76. [17112803 ]
- Li GX, Pei QL, Gao Y, Liu KM, Nie JS, Han G, Qiu YL, Zhang WP: Protective effects of hepatocellular canalicular conjugate export pump (Mrp2) on sodium arsenite-induced hepatic dysfunction in rats. Exp Toxicol Pathol. 2007 Aug;58(6):447-53. Epub 2007 Apr 30. [17467962 ]
- General Function:
- Syntaxin binding
- Specific Function:
- Non-receptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1 and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuromuscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis and modulates the endocytosis of activated B-cell receptor complexes. Other substrates which are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor down-regulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress and thereby mediates mitochondrial dysfunction and cell death. ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA-damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52 or WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates the caspase CASP9 on 'Tyr-153' and regulates its processing in the apoptotic response to DNA damage. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of Vaccinia virus, Tir (translocated intimin receptor) of pathogenic E.coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of H.pylori, or AnkA (ankyrin repeat-containing protein A) of A.phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1.
- Gene Name:
- ABL1
- Uniprot ID:
- P00519
- Molecular Weight:
- 122871.435 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- General Function:
- Tau-protein kinase activity
- Specific Function:
- Catalytic subunit of AMP-activated protein kinase (AMPK), an energy sensor protein kinase that plays a key role in regulating cellular energy metabolism. In response to reduction of intracellular ATP levels, AMPK activates energy-producing pathways and inhibits energy-consuming processes: inhibits protein, carbohydrate and lipid biosynthesis, as well as cell growth and proliferation. AMPK acts via direct phosphorylation of metabolic enzymes, and by longer-term effects via phosphorylation of transcription regulators. Also acts as a regulator of cellular polarity by remodeling the actin cytoskeleton; probably by indirectly activating myosin. Regulates lipid synthesis by phosphorylating and inactivating lipid metabolic enzymes such as ACACA, ACACB, GYS1, HMGCR and LIPE; regulates fatty acid and cholesterol synthesis by phosphorylating acetyl-CoA carboxylase (ACACA and ACACB) and hormone-sensitive lipase (LIPE) enzymes, respectively. Regulates insulin-signaling and glycolysis by phosphorylating IRS1, PFKFB2 and PFKFB3. AMPK stimulates glucose uptake in muscle by increasing the translocation of the glucose transporter SLC2A4/GLUT4 to the plasma membrane, possibly by mediating phosphorylation of TBC1D4/AS160. Regulates transcription and chromatin structure by phosphorylating transcription regulators involved in energy metabolism such as CRTC2/TORC2, FOXO3, histone H2B, HDAC5, MEF2C, MLXIPL/ChREBP, EP300, HNF4A, p53/TP53, SREBF1, SREBF2 and PPARGC1A. Acts as a key regulator of glucose homeostasis in liver by phosphorylating CRTC2/TORC2, leading to CRTC2/TORC2 sequestration in the cytoplasm. In response to stress, phosphorylates 'Ser-36' of histone H2B (H2BS36ph), leading to promote transcription. Acts as a key regulator of cell growth and proliferation by phosphorylating TSC2, RPTOR and ATG1/ULK1: in response to nutrient limitation, negatively regulates the mTORC1 complex by phosphorylating RPTOR component of the mTORC1 complex and by phosphorylating and activating TSC2. In response to nutrient limitation, promotes autophagy by phosphorylating and activating ATG1/ULK1. AMPK also acts as a regulator of circadian rhythm by mediating phosphorylation of CRY1, leading to destabilize it. May regulate the Wnt signaling pathway by phosphorylating CTNNB1, leading to stabilize it. Also has tau-protein kinase activity: in response to amyloid beta A4 protein (APP) exposure, activated by CAMKK2, leading to phosphorylation of MAPT/TAU; however the relevance of such data remains unclear in vivo. Also phosphorylates CFTR, EEF2K, KLC1, NOS3 and SLC12A1.
- Gene Name:
- PRKAA1
- Uniprot ID:
- Q13131
- Molecular Weight:
- 64008.64 Da
References
- General Function:
- Zinc ion binding
- Specific Function:
- ATP-dependent protease which is essential for axonal development.
- Gene Name:
- AFG3L2
- Uniprot ID:
- Q9Y4W6
- Molecular Weight:
- 88583.03 Da
References
- General Function:
- Transmembrane receptor protein tyrosine kinase activity
- Specific Function:
- Neuronal orphan receptor tyrosine kinase that is essentially and transiently expressed in specific regions of the central and peripheral nervous systems and plays an important role in the genesis and differentiation of the nervous system. Transduces signals from ligands at the cell surface, through specific activation of the mitogen-activated protein kinase (MAPK) pathway. Phosphorylates almost exclusively at the first tyrosine of the Y-x-x-x-Y-Y motif. Following activation by ligand, ALK induces tyrosine phosphorylation of CBL, FRS2, IRS1 and SHC1, as well as of the MAP kinases MAPK1/ERK2 and MAPK3/ERK1. Acts as a receptor for ligands pleiotrophin (PTN), a secreted growth factor, and midkine (MDK), a PTN-related factor, thus participating in PTN and MDK signal transduction. PTN-binding induces MAPK pathway activation, which is important for the anti-apoptotic signaling of PTN and regulation of cell proliferation. MDK-binding induces phosphorylation of the ALK target insulin receptor substrate (IRS1), activates mitogen-activated protein kinases (MAPKs) and PI3-kinase, resulting also in cell proliferation induction. Drives NF-kappa-B activation, probably through IRS1 and the activation of the AKT serine/threonine kinase. Recruitment of IRS1 to activated ALK and the activation of NF-kappa-B are essential for the autocrine growth and survival signaling of MDK.
- Gene Name:
- ALK
- Uniprot ID:
- Q9UM73
- Molecular Weight:
- 176440.535 Da
References
- General Function:
- Transporter activity
- Specific Function:
- ATPase required for the post-translational delivery of tail-anchored (TA) proteins to the endoplasmic reticulum. Recognizes and selectively binds the transmembrane domain of TA proteins in the cytosol. This complex then targets to the endoplasmic reticulum by membrane-bound receptors, where the tail-anchored protein is released for insertion. This process is regulated by ATP binding and hydrolysis. ATP binding drives the homodimer towards the closed dimer state, facilitating recognition of newly synthesized TA membrane proteins. ATP hydrolysis is required for insertion. Subsequently, the homodimer reverts towards the open dimer state, lowering its affinity for the membrane-bound receptor, and returning it to the cytosol to initiate a new round of targeting (By similarity). May be involved in insulin signaling.
- Gene Name:
- ASNA1
- Uniprot ID:
- O43681
- Molecular Weight:
- 38792.445 Da
References
- General Function:
- Atp binding
- Specific Function:
- Important for maintaining normal body temperature during fasting and for energy homeostasis. Essential for energy expenditure under ketogenic conditions (By similarity). Converts acetate to acetyl-CoA so that it can be used for oxidation through the tricarboxylic cycle to produce ATP and CO(2).
- Gene Name:
- ACSS1
- Uniprot ID:
- Q9NUB1
- Molecular Weight:
- 74856.1 Da
References
- General Function:
- Ww domain binding
- Specific Function:
- Non-receptor tyrosine-protein and serine/threonine-protein kinase that is implicated in cell spreading and migration, cell survival, cell growth and proliferation. Transduces extracellular signals to cytosolic and nuclear effectors. Phosphorylates AKT1, AR, MCF2, WASL and WWOX. Implicated in trafficking and clathrin-mediated endocytosis through binding to epidermal growth factor receptor (EGFR) and clathrin. Binds to both poly- and mono-ubiquitin and regulates ligand-induced degradation of EGFR, thereby contributing to the accumulation of EGFR at the limiting membrane of early endosomes. Downstream effector of CDC42 which mediates CDC42-dependent cell migration via phosphorylation of BCAR1. May be involved both in adult synaptic function and plasticity and in brain development. Activates AKT1 by phosphorylating it on 'Tyr-176'. Phosphorylates AR on 'Tyr-267' and 'Tyr-363' thereby promoting its recruitment to androgen-responsive enhancers (AREs). Phosphorylates WWOX on 'Tyr-287'. Phosphorylates MCF2, thereby enhancing its activity as a guanine nucleotide exchange factor (GEF) toward Rho family proteins. Contributes to the control of AXL receptor levels. Confers metastatic properties on cancer cells and promotes tumor growth by negatively regulating tumor suppressor such as WWOX and positively regulating pro-survival factors such as AKT1 and AR.
- Gene Name:
- TNK2
- Uniprot ID:
- Q07912
- Molecular Weight:
- 114567.605 Da
References
- General Function:
- Transmembrane receptor protein serine/threonine kinase activity
- Specific Function:
- On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. Receptor for activin. May be involved for left-right pattern formation during embryogenesis (By similarity).
- Gene Name:
- ACVR1
- Uniprot ID:
- Q04771
- Molecular Weight:
- 57152.41 Da
References
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Transmembrane serine/threonine kinase activin type-1 receptor forming an activin receptor complex with activin receptor type-2 (ACVR2A or ACVR2B). Transduces the activin signal from the cell surface to the cytoplasm and is thus regulating a many physiological and pathological processes including neuronal differentiation and neuronal survival, hair follicle development and cycling, FSH production by the pituitary gland, wound healing, extracellular matrix production, immunosuppression and carcinogenesis. Activin is also thought to have a paracrine or autocrine role in follicular development in the ovary. Within the receptor complex, type-2 receptors (ACVR2A and/or ACVR2B) act as a primary activin receptors whereas the type-1 receptors like ACVR1B act as downstream transducers of activin signals. Activin binds to type-2 receptor at the plasma membrane and activates its serine-threonine kinase. The activated receptor type-2 then phosphorylates and activates the type-1 receptor such as ACVR1B. Once activated, the type-1 receptor binds and phosphorylates the SMAD proteins SMAD2 and SMAD3, on serine residues of the C-terminal tail. Soon after their association with the activin receptor and subsequent phosphorylation, SMAD2 and SMAD3 are released into the cytoplasm where they interact with the common partner SMAD4. This SMAD complex translocates into the nucleus where it mediates activin-induced transcription. Inhibitory SMAD7, which is recruited to ACVR1B through FKBP1A, can prevent the association of SMAD2 and SMAD3 with the activin receptor complex, thereby blocking the activin signal. Activin signal transduction is also antagonized by the binding to the receptor of inhibin-B via the IGSF1 inhibin coreceptor. ACVR1B also phosphorylates TDP2.
- Gene Name:
- ACVR1B
- Uniprot ID:
- P36896
- Molecular Weight:
- 56806.05 Da
References
- General Function:
- Metal ion binding
- Specific Function:
- Catalyzes the formation of the signaling molecule cAMP in response to G-protein signaling. Mediates responses to increased cellular Ca(2+)/calmodulin levels (By similarity). May be involved in regulatory processes in the central nervous system. May play a role in memory and learning. Plays a role in the regulation of the circadian rhythm of daytime contrast sensitivity probably by modulating the rhythmic synthesis of cyclic AMP in the retina (By similarity).
- Gene Name:
- ADCY1
- Uniprot ID:
- Q08828
- Molecular Weight:
- 123438.85 Da
References
- General Function:
- Transforming growth factor beta receptor activity, type ii
- Specific Function:
- On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. Receptor for anti-Muellerian hormone.
- Gene Name:
- AMHR2
- Uniprot ID:
- Q16671
- Molecular Weight:
- 62749.02 Da
References
- General Function:
- Toxic substance binding
- Specific Function:
- Is indirectly involved in the control of blood pressure.
- Gene Name:
- ASS1
- Uniprot ID:
- P00966
- Molecular Weight:
- 46530.055 Da
References
- General Function:
- Protein homodimerization activity
- Specific Function:
- Not Available
- Gene Name:
- ASNS
- Uniprot ID:
- P08243
- Molecular Weight:
- 64369.39 Da
References
- General Function:
- Protein kinase activity
- Specific Function:
- Specifically phosphorylates the agonist-occupied form of the beta-adrenergic and closely related receptors, probably inducing a desensitization of them. Key regulator of LPAR1 signaling. Competes with RALA for binding to LPAR1 thus affecting the signaling properties of the receptor. Desensitizes LPAR1 and LPAR2 in a phosphorylation-independent manner.
- Gene Name:
- ADRBK1
- Uniprot ID:
- P25098
- Molecular Weight:
- 79572.96 Da
References
- General Function:
- Protein kinase activity
- Specific Function:
- Specifically phosphorylates the agonist-occupied form of the beta-adrenergic and closely related receptors.
- Gene Name:
- ADRBK2
- Uniprot ID:
- P35626
- Molecular Weight:
- 79709.085 Da
References
- General Function:
- Transporter activity
- Specific Function:
- Involved in the ATP-dependent secretion of bile salts into the canaliculus of hepatocytes.
- Gene Name:
- ABCB11
- Uniprot ID:
- O95342
- Molecular Weight:
- 146405.83 Da
References
- Snow KL, Moseley RH: Effect of thiazolidinediones on bile acid transport in rat liver. Life Sci. 2007 Jan 30;80(8):732-40. Epub 2006 Nov 10. [17126857 ]
- Suchy FJ, Ananthanarayanan M: Bile salt excretory pump: biology and pathobiology. J Pediatr Gastroenterol Nutr. 2006 Jul;43 Suppl 1:S10-6. [16819395 ]
- General Function:
- Protein serine/threonine kinase activity
- Specific Function:
- Serine/threonine-protein kinase that acts like an antiapoptotic protein that counters TRAIL/TNFSF10-induced apoptosis by inducing phosphorylation of BIRC5 at 'Thr-34'.
- Gene Name:
- CDK15
- Uniprot ID:
- Q96Q40
- Molecular Weight:
- 49022.64 Da
References
- General Function:
- Nucleotide binding
- Specific Function:
- May have a critical role in the maintenance of a constant composition of intracellular purine/pyrimidine nucleotides in cooperation with other nucleotidases. Preferentially hydrolyzes inosine 5'-monophosphate (IMP) and other purine nucleotides.
- Gene Name:
- NT5C2
- Uniprot ID:
- P49902
- Molecular Weight:
- 64969.2 Da
References
- General Function:
- Long-chain fatty acid-coa ligase activity
- Specific Function:
- Activation of long-chain fatty acids for both synthesis of cellular lipids, and degradation via beta-oxidation. Preferentially uses palmitoleate, oleate and linoleate.
- Gene Name:
- ACSL1
- Uniprot ID:
- P33121
- Molecular Weight:
- 77942.685 Da
References
- General Function:
- Transporter activity
- Specific Function:
- Mediates export of organic anions and drugs from the cytoplasm. Mediates ATP-dependent transport of glutathione and glutathione conjugates, leukotriene C4, estradiol-17-beta-o-glucuronide, methotrexate, antiviral drugs and other xenobiotics. Confers resistance to anticancer drugs. Hydrolyzes ATP with low efficiency.
- Gene Name:
- ABCC1
- Uniprot ID:
- P33527
- Molecular Weight:
- 171589.5 Da
References
- Zhao Q, Chang XB: Mutation of the aromatic amino acid interacting with adenine moiety of ATP to a polar residue alters the properties of multidrug resistance protein 1. J Biol Chem. 2004 Nov 19;279(47):48505-12. Epub 2004 Sep 7. [15355964 ]
- Yang R, McBride A, Hou YX, Goldberg A, Chang XB: Nucleotide dissociation from NBD1 promotes solute transport by MRP1. Biochim Biophys Acta. 2005 Mar 1;1668(2):248-61. [15737336 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Regulatory subunit of the dimeric UBA3-NAE1 E1 enzyme. E1 activates NEDD8 by first adenylating its C-terminal glycine residue with ATP, thereafter linking this residue to the side chain of the catalytic cysteine, yielding a NEDD8-UBA3 thioester and free AMP. E1 finally transfers NEDD8 to the catalytic cysteine of UBE2M. Necessary for cell cycle progression through the S-M checkpoint. Overexpression of NAE1 causes apoptosis through deregulation of NEDD8 conjugation.
- Gene Name:
- NAE1
- Uniprot ID:
- Q13564
- Molecular Weight:
- 60245.795 Da
References
- General Function:
- Receptor signaling protein activity
- Specific Function:
- Involved in the transduction of mitogenic signals from the cell membrane to the nucleus. May also regulate the TOR signaling cascade.Isoform 2: Serves as a positive regulator of myogenic differentiation by inducing cell cycle arrest, the expression of myogenin and other muscle-specific proteins, and myotube formation.
- Gene Name:
- ARAF
- Uniprot ID:
- P10398
- Molecular Weight:
- 67584.825 Da
References
- General Function:
- Transmembrane receptor protein serine/threonine kinase activity
- Specific Function:
- Type I receptor for TGF-beta family ligands BMP9/GDF2 and BMP10 and important regulator of normal blood vessel development. On ligand binding, forms a receptor complex consisting of two type II and two type I transmembrane serine/threonine kinases. Type II receptors phosphorylate and activate type I receptors which autophosphorylate, then bind and activate SMAD transcriptional regulators. May bind activin as well.
- Gene Name:
- ACVRL1
- Uniprot ID:
- P37023
- Molecular Weight:
- 56124.03 Da
References
- General Function:
- Adenine transmembrane transporter activity
- Specific Function:
- Catalyzes the exchange of cytoplasmic ADP with mitochondrial ATP across the mitochondrial inner membrane.
- Gene Name:
- SLC25A4
- Uniprot ID:
- P12235
- Molecular Weight:
- 33064.265 Da
References
- Walther T, Tschope C, Sterner-Kock A, Westermann D, Heringer-Walther S, Riad A, Nikolic A, Wang Y, Ebermann L, Siems WE, Bader M, Shakibaei M, Schultheiss HP, Dorner A: Accelerated mitochondrial adenosine diphosphate/adenosine triphosphate transport improves hypertension-induced heart disease. Circulation. 2007 Jan 23;115(3):333-44. Epub 2007 Jan 8. [17210842 ]
- General Function:
- Pdz domain binding
- Specific Function:
- Involved in the transport of chloride ions. May regulate bicarbonate secretion and salvage in epithelial cells by regulating the SLC4A7 transporter. Can inhibit the chloride channel activity of ANO1. Plays a role in the chloride and bicarbonate homeostasis during sperm epididymal maturation and capacitation.
- Gene Name:
- CFTR
- Uniprot ID:
- P13569
- Molecular Weight:
- 168139.895 Da
References
- Berger AL, Ikuma M, Welsh MJ: Normal gating of CFTR requires ATP binding to both nucleotide-binding domains and hydrolysis at the second nucleotide-binding domain. Proc Natl Acad Sci U S A. 2005 Jan 11;102(2):455-60. Epub 2004 Dec 27. [15623556 ]
35. Heat shock 70 kDa protein 1A/1B (Protein Group)
- General Function:
- Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, folding and transport of newly synthesized polypeptides, activation of proteolysis of misfolded proteins and the formation and dissociation of protein complexes. Plays a pivotal role in the protein quality control system, ensuring the correct folding of proteins, the re-folding of misfolded proteins and controlling the targeting of proteins for subsequent degradation. This is achieved through cycles of ATP binding, ATP hydrolysis and ADP release, mediated by co-chaperones. The co-chaperones have been shown to not only regulate different steps of the ATPase cycle, but they also have an individual specificity such that one co-chaperone may promote folding of a substrate while another may promote degradation. The affinity for polypeptides is regulated by its nucleotide bound state. In the ATP-bound form, it has a low affinity for substrate proteins. However, upon hydrolysis of the ATP to ADP, it undergoes a conformational change that increases its affinity for substrate proteins. It goes through repeated cycles of ATP hydrolysis and nucleotide exchange, which permits cycles of substrate binding and release. The co-chaperones are of three types: J-domain co-chaperones such as HSP40s (stimulate ATPase hydrolysis by HSP70), the nucleotide exchange factors (NEF) such as BAG1/2/3 (facilitate conversion of HSP70 from the ADP-bound to the ATP-bound state thereby promoting substrate release), and the TPR domain chaperones such as HOPX and STUB1 (PubMed:24012426, PubMed:26865365, PubMed:24318877). Maintains protein homeostasis during cellular stress through two opposing mechanisms: protein refolding and degradation. Its acetylation/deacetylation state determines whether it functions in protein refolding or protein degradation by controlling the competitive binding of co-chaperones HOPX and STUB1. During the early stress response, the acetylated form binds to HOPX which assists in chaperone-mediated protein refolding, thereafter, it is deacetylated and binds to ubiquitin ligase STUB1 that promotes ubiquitin-mediated protein degradation (PubMed:27708256). Regulates centrosome integrity during mitosis, and is required for the maintenance of a functional mitotic centrosome that supports the assembly of a bipolar mitotic spindle (PubMed:27137183). Enhances STUB1-mediated SMAD3 ubiquitination and degradation and facilitates STUB1-mediated inhibition of TGF-beta signaling (PubMed:24613385). Essential for STUB1-mediated ubiquitination and degradation of FOXP3 in regulatory T-cells (Treg) during inflammation (PubMed:23973223). Negatively regulates heat shock-induced HSF1 transcriptional activity during the attenuation and recovery phase period of the heat shock response (PubMed:9499401).
- Specific Function:
- Atp binding
- Included Proteins:
- P0DMV8 , P0DMV9
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 0.5 uM | Not Available | BindingDB 2 |
References
- Williamson DS, Borgognoni J, Clay A, Daniels Z, Dokurno P, Drysdale MJ, Foloppe N, Francis GL, Graham CJ, Howes R, Macias AT, Murray JB, Parsons R, Shaw T, Surgenor AE, Terry L, Wang Y, Wood M, Massey AJ: Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J Med Chem. 2009 Mar 26;52(6):1510-3. doi: 10.1021/jm801627a. [19256508 ]
- General Function:
- Transporter activity
- Specific Function:
- Isoform 1: May participate directly in the active transport of drugs into subcellular organelles or influence drug distribution indirectly. Transports glutathione conjugates as leukotriene-c4 (LTC4) and N-ethylmaleimide S-glutathione (NEM-GS).Isoform 2: Inhibits TNF-alpha-mediated apoptosis through blocking one or more caspases.
- Gene Name:
- ABCC6
- Uniprot ID:
- O95255
- Molecular Weight:
- 164904.81 Da
References
- Ilias A, Urban Z, Seidl TL, Le Saux O, Sinko E, Boyd CD, Sarkadi B, Varadi A: Loss of ATP-dependent transport activity in pseudoxanthoma elasticum-associated mutants of human ABCC6 (MRP6). J Biol Chem. 2002 May 10;277(19):16860-7. Epub 2002 Mar 5. [11880368 ]
- General Function:
- Purinergic nucleotide receptor activity
- Specific Function:
- Receptor for ATP that acts as a ligand-gated ion channel.
- Gene Name:
- P2RX3
- Uniprot ID:
- P56373
- Molecular Weight:
- 44288.65 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.0146 uM | Not Available | BindingDB 2 |
References
- Jarvis MF, Bianchi B, Uchic JT, Cartmell J, Lee CH, Williams M, Faltynek C: [3H]A-317491, a novel high-affinity non-nucleotide antagonist that specifically labels human P2X2/3 and P2X3 receptors. J Pharmacol Exp Ther. 2004 Jul;310(1):407-16. Epub 2004 Mar 15. [15024037 ]
- General Function:
- Scaffold protein binding
- Specific Function:
- Receptor for extracellular adenine nucleotides such as ATP and ADP. In platelets binding to ADP leads to mobilization of intracellular calcium ions via activation of phospholipase C, a change in platelet shape, and probably to platelet aggregation.
- Gene Name:
- P2RY1
- Uniprot ID:
- P47900
- Molecular Weight:
- 42071.08 Da
References
- Fischer W, Wirkner K, Weber M, Eberts C, Koles L, Reinhardt R, Franke H, Allgaier C, Gillen C, Illes P: Characterization of P2X3, P2Y1 and P2Y4 receptors in cultured HEK293-hP2X3 cells and their inhibition by ethanol and trichloroethanol. J Neurochem. 2003 May;85(3):779-90. [12694404 ]
- General Function:
- Receptor activity
- Specific Function:
- Receptor for ATP and ADP coupled to G-proteins that activate both phosphatidylinositol-calcium and adenylyl cyclase second messenger systems. Not activated by UTP or UDP.
- Gene Name:
- P2RY11
- Uniprot ID:
- Q96G91
- Molecular Weight:
- 40344.755 Da
References
- Xiao Z, Yang M, Lv Q, Wang W, Deng M, Liu X, He Q, Chen X, Chen M, Fang L, Xie X, Hu J: P2Y11 impairs cell proliferation by induction of cell cycle arrest and sensitizes endothelial cells to cisplatin-induced cell death. J Cell Biochem. 2011 Sep;112(9):2257-65. doi: 10.1002/jcb.23144. [21503959 ]
- General Function:
- Utp-activated nucleotide receptor activity
- Specific Function:
- Receptor for UTP and UDP coupled to G-proteins that activate a phosphatidylinositol-calcium second messenger system. Not activated by ATP or ADP.
- Gene Name:
- P2RY4
- Uniprot ID:
- P51582
- Molecular Weight:
- 40962.49 Da
References
- Fischer W, Wirkner K, Weber M, Eberts C, Koles L, Reinhardt R, Franke H, Allgaier C, Gillen C, Illes P: Characterization of P2X3, P2Y1 and P2Y4 receptors in cultured HEK293-hP2X3 cells and their inhibition by ethanol and trichloroethanol. J Neurochem. 2003 May;85(3):779-90. [12694404 ]
- General Function:
- Protein serine/threonine kinase activity
- Specific Function:
- Phosphoinositide-3-kinase (PI3K) that phosphorylates PtdIns (Phosphatidylinositol), PtdIns4P (Phosphatidylinositol 4-phosphate) and PtdIns(4,5)P2 (Phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Participates in cellular signaling in response to various growth factors. Involved in the activation of AKT1 upon stimulation by receptor tyrosine kinases ligands such as EGF, insulin, IGF1, VEGFA and PDGF. Involved in signaling via insulin-receptor substrate (IRS) proteins. Essential in endothelial cell migration during vascular development through VEGFA signaling, possibly by regulating RhoA activity. Required for lymphatic vasculature development, possibly by binding to RAS and by activation by EGF and FGF2, but not by PDGF. Regulates invadopodia formation in breast cancer cells through the PDPK1-AKT1 pathway. Participates in cardiomyogenesis in embryonic stem cells through a AKT1 pathway. Participates in vasculogenesis in embryonic stem cells through PDK1 and protein kinase C pathway. Has also serine-protein kinase activity: phosphorylates PIK3R1 (p85alpha regulatory subunit), EIF4EBP1 and HRAS.
- Gene Name:
- PIK3CA
- Uniprot ID:
- P42336
- Molecular Weight:
- 124283.025 Da
References
- Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box G, Chuckowree IS, Clarke PA, Depledge P, Eccles SA, Friedman LS, Hayes A, Hancox TC, Kugendradas A, Lensun L, Moore P, Olivero AG, Pang J, Patel S, Pergl-Wilson GH, Raynaud FI, Robson A, Saghir N, Salphati L, Sohal S, Ultsch MH, Valenti M, Wallweber HJ, Wan NC, Wiesmann C, Workman P, Zhyvoloup A, Zvelebil MJ, Shuttleworth SJ: The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-t hieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer . J Med Chem. 2008 Sep 25;51(18):5522-32. doi: 10.1021/jm800295d. [18754654 ]
- General Function:
- Protein serine/threonine/tyrosine kinase activity
- Specific Function:
- AKT1 is one of 3 closely related serine/threonine-protein kinases (AKT1, AKT2 and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth and angiogenesis. This is mediated through serine and/or threonine phosphorylation of a range of downstream substrates. Over 100 substrate candidates have been reported so far, but for most of them, no isoform specificity has been reported. AKT is responsible of the regulation of glucose uptake by mediating insulin-induced translocation of the SLC2A4/GLUT4 glucose transporter to the cell surface. Phosphorylation of PTPN1 at 'Ser-50' negatively modulates its phosphatase activity preventing dephosphorylation of the insulin receptor and the attenuation of insulin signaling. Phosphorylation of TBC1D4 triggers the binding of this effector to inhibitory 14-3-3 proteins, which is required for insulin-stimulated glucose transport. AKT regulates also the storage of glucose in the form of glycogen by phosphorylating GSK3A at 'Ser-21' and GSK3B at 'Ser-9', resulting in inhibition of its kinase activity. Phosphorylation of GSK3 isoforms by AKT is also thought to be one mechanism by which cell proliferation is driven. AKT regulates also cell survival via the phosphorylation of MAP3K5 (apoptosis signal-related kinase). Phosphorylation of 'Ser-83' decreases MAP3K5 kinase activity stimulated by oxidative stress and thereby prevents apoptosis. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 at 'Ser-939' and 'Thr-1462', thereby activating mTORC1 signaling and leading to both phosphorylation of 4E-BP1 and in activation of RPS6KB1. AKT is involved in the phosphorylation of members of the FOXO factors (Forkhead family of transcription factors), leading to binding of 14-3-3 proteins and cytoplasmic localization. In particular, FOXO1 is phosphorylated at 'Thr-24', 'Ser-256' and 'Ser-319'. FOXO3 and FOXO4 are phosphorylated on equivalent sites. AKT has an important role in the regulation of NF-kappa-B-dependent gene transcription and positively regulates the activity of CREB1 (cyclic AMP (cAMP)-response element binding protein). The phosphorylation of CREB1 induces the binding of accessory proteins that are necessary for the transcription of pro-survival genes such as BCL2 and MCL1. AKT phosphorylates 'Ser-454' on ATP citrate lyase (ACLY), thereby potentially regulating ACLY activity and fatty acid synthesis. Activates the 3B isoform of cyclic nucleotide phosphodiesterase (PDE3B) via phosphorylation of 'Ser-273', resulting in reduced cyclic AMP levels and inhibition of lipolysis. Phosphorylates PIKFYVE on 'Ser-318', which results in increased PI(3)P-5 activity. The Rho GTPase-activating protein DLC1 is another substrate and its phosphorylation is implicated in the regulation cell proliferation and cell growth. AKT plays a role as key modulator of the AKT-mTOR signaling pathway controlling the tempo of the process of newborn neurons integration during adult neurogenesis, including correct neuron positioning, dendritic development and synapse formation. Signals downstream of phosphatidylinositol 3-kinase (PI(3)K) to mediate the effects of various growth factors such as platelet-derived growth factor (PDGF), epidermal growth factor (EGF), insulin and insulin-like growth factor I (IGF-I). AKT mediates the antiapoptotic effects of IGF-I. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. May be involved in the regulation of the placental development. Phosphorylates STK4/MST1 at 'Thr-120' and 'Thr-387' leading to inhibition of its: kinase activity, nuclear translocation, autophosphorylation and ability to phosphorylate FOXO3. Phosphorylates STK3/MST2 at 'Thr-117' and 'Thr-384' leading to inhibition of its: cleavage, kinase activity, autophosphorylation at Thr-180, binding to RASSF1 and nuclear translocation. Phosphorylates SRPK2 and enhances its kinase activity towards SRSF2 and ACIN1 and promotes its nuclear translocation. Phosphorylates RAF1 at 'Ser-259' and negatively regulates its activity. Phosphorylation of BAD stimulates its pro-apoptotic activity. Phosphorylates KAT6A at 'Thr-369' and this phosphorylation inhibits the interaction of KAT6A with PML and negatively regulates its acetylation activity towards p53/TP53.AKT1-specific substrates have been recently identified, including palladin (PALLD), which phosphorylation modulates cytoskeletal organization and cell motility; prohibitin (PHB), playing an important role in cell metabolism and proliferation; and CDKN1A, for which phosphorylation at 'Thr-145' induces its release from CDK2 and cytoplasmic relocalization. These recent findings indicate that the AKT1 isoform has a more specific role in cell motility and proliferation. Phosphorylates CLK2 thereby controlling cell survival to ionizing radiation.
- Gene Name:
- AKT1
- Uniprot ID:
- P31749
- Molecular Weight:
- 55686.035 Da
References
- Van Meter TE, Broaddus WC, Cash D, Fillmore H: Cotreatment with a novel phosphoinositide analogue inhibitor and carmustine enhances chemotherapeutic efficacy by attenuating AKT activity in gliomas. Cancer. 2006 Nov 15;107(10):2446-54. [17041888 ]