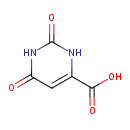
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-0udi-1982000000-b20f2ec9a2f1acc84ae1 | 2014-06-16 | View Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-0zfr-2693000000-07bf11b5177412647d9b | 2014-06-16 | View Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0udi-1982000000-b20f2ec9a2f1acc84ae1 | 2017-09-12 | View Spectrum |
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-0zfr-2693000000-07bf11b5177412647d9b | 2017-09-12 | View Spectrum |
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-0udi-1972000000-c04fb7381d3ca07c9b0a | 2017-09-12 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-06ri-6900000000-4ba5fde1f8f62c4a2ba2 | 2016-09-22 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0229-9850000000-8164142820eb5162e753 | 2017-10-06 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum |
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-03di-0900000000-81d73aed73c250fff3ae | 2012-07-24 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0006-9300000000-ecb08d1854a2789480d1 | 2012-07-24 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0006-9100000000-15748a4c13636c42f4f9 | 2012-07-24 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-02ti-4900000000-6fd6312d81f94faaaf11 | 2012-08-31 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-03di-0900000000-1b7abf52e4a01cb85541 | 2012-08-31 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-03di-0900000000-1b7abf52e4a01cb85541 | 2017-09-14 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-03di-0900000000-3dff3caa5c10aff8ce46 | 2017-09-14 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-02ti-4900000000-6fd6312d81f94faaaf11 | 2017-09-14 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-004i-9000000000-351f585d58b38f6132de | 2021-09-20 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0a4r-0900000000-4e46209ca236b470bf7f | 2021-09-20 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-014i-9300000000-aa9cd31d715d134ee8dd | 2021-09-20 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-014i-9000000000-cdd01e740b6f9d1a909a | 2021-09-20 | View Spectrum |
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-03di-1900000000-baf39ad8ab6f08fb1a7b | 2021-09-20 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0bt9-0900000000-1ad59feda583f2247b21 | 2015-04-24 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-2900000000-0d0d27215e9f39a2a15b | 2015-04-24 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-918a38777f5d4e049697 | 2015-04-24 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-d7bbee7f2dc7bb0c6a83 | 2015-04-25 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dl-7900000000-303fe3e06e98f518a2ee | 2015-04-25 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-b2c02975bbba65c2f1c6 | 2015-04-25 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0bt9-0900000000-1ad59feda583f2247b21 | 2015-05-27 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-2900000000-0d0d27215e9f39a2a15b | 2015-05-27 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9100000000-918a38777f5d4e049697 | 2015-05-27 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-d7bbee7f2dc7bb0c6a83 | 2015-05-27 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dl-7900000000-303fe3e06e98f518a2ee | 2015-05-27 | View Spectrum |
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-b2c02975bbba65c2f1c6 | 2015-05-27 | View Spectrum |
| MS | Mass Spectrum (Electron Ionization) | splash10-066r-9300000000-e5c3ef3304f93a38628c | 2014-09-20 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | 2012-12-04 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 90 MHz, DMSO-d6, experimental) | Not Available | 2014-09-20 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-16 | View Spectrum |
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | 2012-12-04 | View Spectrum |
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | 2012-12-05 | View Spectrum |