S-Adenosylhomocysteine (T3D4455)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2014-08-29 06:51:24 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2018-03-21 17:46:15 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D4455 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | S-Adenosylhomocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | S-Adenosyl-L-homocysteine (SAH) is formed by the demethylation of S-adenosyl-L-methionine. S-Adenosylhomocysteine (AdoHcy or SAH) is also the immediate precursor of all of the homocysteine produced in the body. The reaction is catalyzed by S-adenosylhomocysteine hydrolase and is reversible with the equilibrium favoring formation of SAH. In vivo, the reaction is driven in the direction of homocysteine formation by the action of the enzyme adenosine deaminase, which converts the second product of the S-adenosylhomocysteine hydrolase reaction, adenosine, to inosine. Except for methyl transfer from betaine and from methylcobalamin in the methionine synthase reaction, SAH is the product of all methylation reactions that involve S-adenosylmethionine (SAM) as the methyl donor. Methylation is significant in epigenetic regulation of protein expression via DNA and histone methylation. The inhibition of these SAM-mediated processes by SAH is a proven mechanism for metabolic alteration. Because the conversion of SAH to homocysteine is reversible, with the equilibrium favoring the formation of SAH, increases in plasma homocysteine are accompanied by an elevation of SAH in most cases. Disturbances in the transmethylation pathway indicated by abnormal SAH, SAM or their ratio have been reported in many neurodegenerative diseases, such as dementia, depression or Parkinson's disease. (PMID: 18065573 , 17892439). Therefore, when present in sufficiently high levels, S-adenosylhomocysteine can act as an immunotoxin, and a metabotoxin. An immunotoxin disrupts, limits the function, or destroys immune cells. A metabotoxin is an endogenous metabolite that causes adverse health effects at chronically high levels. Chronically high levels of S-adenosylhomocysteine are associated with S-Adenosylhomocysteine (SAH) Hydrolase Deficiency and Adenosine Deaminase Deficiency. S-adenosylhomocysteine forms when there are elevated levels of homocysteine and adenosine. S-adenosyl-L-homocysteine is a potent inhibitor of S-adenosyl-L-methionine dependent methylation reactions. It is toxic to immature lymphocytes and can lead to immunosuppression (PMID: 221926). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
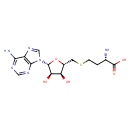
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C14H20N6O5S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 384.411 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 384.122 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 979-92-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl}sulfanyl)butanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | S-adenosyl-L-homocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@](N)(CCSC[C@@]1([H])O[C@@]([H])(N2C=NC3=C(N)N=CN=C23)[C@]([H])(O)[C@]1([H])O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C14H20N6O5S/c15-6(14(23)24)1-2-26-3-7-9(21)10(22)13(25-7)20-5-19-8-11(16)17-4-18-12(8)20/h4-7,9-10,13,21-22H,1-3,15H2,(H,23,24)(H2,16,17,18)/t6-,7+,9+,10+,13+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=ZJUKTBDSGOFHSH-WFMPWKQPSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as gamma butyrolactones. Gamma butyrolactones are compounds containing a gamma butyrolactone moiety, which consists of an aliphatic five-member ring with four carbon atoms, one oxygen atom, and bears a ketone group on the carbon adjacent to the oxygen atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Lactones | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Gamma butyrolactones | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Gamma butyrolactones | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Endogenous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | This is an endogenously produced metabolite found in the human body. It is used in metabolic reactions, catabolic reactions or waste generation. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Chronically high levels of S-adenosylhomocysteine are associated with S-Adenosylhomocysteine (SAH) Hydrolase Deficiency. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB01752 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB00939 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 439155 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL418052 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 388301 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C00021 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 16680 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | ADENOSYL-HOMO-CYS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | SAH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | SAH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Hideaki Yamada, Sakayu Shimizu, Shozo Shiozaki, “Process for producing S-adenosyl-L-homocysteine.” U.S. Patent US4605625, issued March, 1978. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Zinc ion binding
- Specific Function:
- Methylates CpG residues. Preferentially methylates hemimethylated DNA. Associates with DNA replication sites in S phase maintaining the methylation pattern in the newly synthesized strand, that is essential for epigenetic inheritance. Associates with chromatin during G2 and M phases to maintain DNA methylation independently of replication. It is responsible for maintaining methylation patterns established in development. DNA methylation is coordinated with methylation of histones. Mediates transcriptional repression by direct binding to HDAC2. In association with DNMT3B and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9.
- Gene Name:
- DNMT1
- Uniprot ID:
- P26358
- Molecular Weight:
- 183163.635 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 0.8 uM | Not Available | BindingDB 50009672 |
| IC50 | 0.9 uM | Not Available | BindingDB 50009672 |
| IC50 | 0.941 uM | Not Available | BindingDB 50009672 |
| IC50 | 2 uM | Not Available | BindingDB 50009672 |
| IC50 | 4 uM | Not Available | BindingDB 50009672 |
References
- Isakovic L, Saavedra OM, Llewellyn DB, Claridge S, Zhan L, Bernstein N, Vaisburg A, Elowe N, Petschner AJ, Rahil J, Beaulieu N, Gauthier F, MacLeod AR, Delorme D, Besterman JM, Wahhab A: Constrained (l-)-S-adenosyl-l-homocysteine (SAH) analogues as DNA methyltransferase inhibitors. Bioorg Med Chem Lett. 2009 May 15;19(10):2742-6. doi: 10.1016/j.bmcl.2009.03.132. Epub 2009 Mar 28. [19364644 ]
- Baud MG, Leiser T, Haus P, Samlal S, Wong AC, Wood RJ, Petrucci V, Gunaratnam M, Hughes SM, Buluwela L, Turlais F, Neidle S, Meyer-Almes FJ, White AJ, Fuchter MJ: Defining the mechanism of action and enzymatic selectivity of psammaplin A against its epigenetic targets. J Med Chem. 2012 Feb 23;55(4):1731-50. doi: 10.1021/jm2016182. Epub 2012 Feb 9. [22280363 ]
- Hobley G, McKelvie JC, Harmer JE, Howe J, Oyston PC, Roach PL: Development of rationally designed DNA N6 adenine methyltransferase inhibitors. Bioorg Med Chem Lett. 2012 May 1;22(9):3079-82. doi: 10.1016/j.bmcl.2012.03.072. Epub 2012 Mar 27. [22483584 ]
- Saavedra OM, Isakovic L, Llewellyn DB, Zhan L, Bernstein N, Claridge S, Raeppel F, Vaisburg A, Elowe N, Petschner AJ, Rahil J, Beaulieu N, MacLeod AR, Delorme D, Besterman JM, Wahhab A: SAR around (l)-S-adenosyl-l-homocysteine, an inhibitor of human DNA methyltransferase (DNMT) enzymes. Bioorg Med Chem Lett. 2009 May 15;19(10):2747-51. doi: 10.1016/j.bmcl.2009.03.113. Epub 2009 Mar 28. [19362833 ]
- Kuck D, Singh N, Lyko F, Medina-Franco JL: Novel and selective DNA methyltransferase inhibitors: Docking-based virtual screening and experimental evaluation. Bioorg Med Chem. 2010 Jan 15;18(2):822-9. doi: 10.1016/j.bmc.2009.11.050. Epub 2009 Nov 27. [20006515 ]
- Castellano S, Kuck D, Viviano M, Yoo J, Lopez-Vallejo F, Conti P, Tamborini L, Pinto A, Medina-Franco JL, Sbardella G: Synthesis and biochemical evaluation of delta(2)-isoxazoline derivatives as DNA methyltransferase 1 inhibitors. J Med Chem. 2011 Nov 10;54(21):7663-77. doi: 10.1021/jm2010404. Epub 2011 Oct 7. [21958292 ]
- General Function:
- O-methyltransferase activity
- Specific Function:
- Putative O-methyltransferase.
- Gene Name:
- COMTD1
- Uniprot ID:
- Q86VU5
- Molecular Weight:
- 28808.225 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Unmethylated cpg binding
- Specific Function:
- Required for genome-wide de novo methylation and is essential for the establishment of DNA methylation patterns during development. DNA methylation is coordinated with methylation of histones. May preferentially methylates nucleosomal DNA within the nucleosome core region. May function as transcriptional co-repressor by associating with CBX4 and independently of DNA methylation. Seems to be involved in gene silencing (By similarity). In association with DNMT1 and via the recruitment of CTCFL/BORIS, involved in activation of BAG1 gene expression by modulating dimethylation of promoter histone H3 at H3K4 and H3K9. Isoforms 4 and 5 are probably not functional due to the deletion of two conserved methyltransferase motifs. Function as transcriptional corepressor by associating with ZHX1.
- Gene Name:
- DNMT3B
- Uniprot ID:
- Q9UBC3
- Molecular Weight:
- 95750.18 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 0.2 uM | Not Available | BindingDB 50009672 |
| IC50 | 0.25 uM | Not Available | BindingDB 50009672 |
| IC50 | 0.3 uM | Not Available | BindingDB 50009672 |
References
- Isakovic L, Saavedra OM, Llewellyn DB, Claridge S, Zhan L, Bernstein N, Vaisburg A, Elowe N, Petschner AJ, Rahil J, Beaulieu N, Gauthier F, MacLeod AR, Delorme D, Besterman JM, Wahhab A: Constrained (l-)-S-adenosyl-l-homocysteine (SAH) analogues as DNA methyltransferase inhibitors. Bioorg Med Chem Lett. 2009 May 15;19(10):2742-6. doi: 10.1016/j.bmcl.2009.03.132. Epub 2009 Mar 28. [19364644 ]
- Kuck D, Singh N, Lyko F, Medina-Franco JL: Novel and selective DNA methyltransferase inhibitors: Docking-based virtual screening and experimental evaluation. Bioorg Med Chem. 2010 Jan 15;18(2):822-9. doi: 10.1016/j.bmc.2009.11.050. Epub 2009 Nov 27. [20006515 ]
- Saavedra OM, Isakovic L, Llewellyn DB, Zhan L, Bernstein N, Claridge S, Raeppel F, Vaisburg A, Elowe N, Petschner AJ, Rahil J, Beaulieu N, MacLeod AR, Delorme D, Besterman JM, Wahhab A: SAR around (l)-S-adenosyl-l-homocysteine, an inhibitor of human DNA methyltransferase (DNMT) enzymes. Bioorg Med Chem Lett. 2009 May 15;19(10):2747-51. doi: 10.1016/j.bmcl.2009.03.113. Epub 2009 Mar 28. [19362833 ]
- General Function:
- Methyltransferase activity
- Specific Function:
- Not Available
- Gene Name:
- GAMT
- Uniprot ID:
- Q14353
- Molecular Weight:
- 26317.925 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Protein-l-isoaspartate (d-aspartate) o-methyltransferase activity
- Specific Function:
- Catalyzes the methyl esterification of L-isoaspartyl and D-aspartyl residues in peptides and proteins that result from spontaneous decomposition of normal L-aspartyl and L-asparaginyl residues. It plays a role in the repair and/or degradation of damaged proteins. Acts on EIF4EBP2, microtubule-associated protein 2, calreticulin, clathrin light chains a and b, Ubiquitin carboxyl-terminal hydrolase isozyme L1, phosphatidylethanolamine-binding protein 1, stathmin, beta-synuclein and alpha-synuclein.
- Gene Name:
- PCMT1
- Uniprot ID:
- P22061
- Molecular Weight:
- 24636.21 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Transcription factor binding
- Specific Function:
- Histone methyltransferase. Methylates 'Lys-79' of histone H3. Nucleosomes are preferred as substrate compared to free histones. Binds to DNA.
- Gene Name:
- DOT1L
- Uniprot ID:
- Q8TEK3
- Molecular Weight:
- 184851.18 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.16 uM | Not Available | BindingDB 50009672 |
| IC50 | 0.6 uM | Not Available | BindingDB 50009672 |
| Dissociation | 0.15 uM | Not Available | BindingDB 50009672 |
| Dissociation | 0.36 uM | Not Available | BindingDB 50009672 |
References
- Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y: Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J Med Chem. 2012 Sep 27;55(18):8066-74. Epub 2012 Sep 6. [22924785 ]
- Yu W, Smil D, Li F, Tempel W, Fedorov O, Nguyen KT, Bolshan Y, Al-Awar R, Knapp S, Arrowsmith CH, Vedadi M, Brown PJ, Schapira M: Bromo-deaza-SAH: a potent and selective DOT1L inhibitor. Bioorg Med Chem. 2013 Apr 1;21(7):1787-94. doi: 10.1016/j.bmc.2013.01.049. Epub 2013 Jan 30. [23433670 ]
- General Function:
- Phenylethanolamine n-methyltransferase activity
- Specific Function:
- Converts noradrenaline to adrenaline.
- Gene Name:
- PNMT
- Uniprot ID:
- P11086
- Molecular Weight:
- 30854.745 Da
References
- General Function:
- Protein-arginine omega-n asymmetric methyltransferase activity
- Specific Function:
- Arginine methyltransferase that methylates (mono and asymmetric dimethylation) the guanidino nitrogens of arginyl residues present in proteins such as ESR1, histone H2, H3 and H4, PIAS1, HNRNPA1, HNRNPD, NFATC2IP, SUPT5H, TAF15 and EWS. Constitutes the main enzyme that mediates monomethylation and asymmetric dimethylation of histone H4 'Arg-4' (H4R3me1 and H4R3me2a, respectively), a specific tag for epigenetic transcriptional activation. Together with dimethylated PIAS1, represses STAT1 transcriptional activity, in the late phase of interferon gamma (IFN-gamma) signaling. May be involved in the regulation of TAF15 transcriptional activity, act as an activator of estrogen receptor (ER)-mediated transactivation, play a key role in neurite outgrowth and act as a negative regulator of megakaryocytic differentiation, by modulating p38 MAPK pathway. Methylates FOXO1 and retains it in the nucleus increasing its transcriptional activity.
- Gene Name:
- PRMT1
- Uniprot ID:
- Q99873
- Molecular Weight:
- 41515.2 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.4 uM | Not Available | BindingDB 50009672 |
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y: Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J Med Chem. 2012 Sep 27;55(18):8066-74. Epub 2012 Sep 6. [22924785 ]
- General Function:
- Diphthine synthase activity
- Specific Function:
- S-adenosyl-L-methionine-dependent methyltransferase that catalyzes four methylations of the modified target histidine residue in translation elongation factor 2 (EF-2), to form an intermediate called diphthine methyl ester. The four successive methylation reactions represent the second step of diphthamide biosynthesis.
- Gene Name:
- DPH5
- Uniprot ID:
- Q9H2P9
- Molecular Weight:
- 31651.17 Da
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Glycine n-methyltransferase activity
- Specific Function:
- Catalyzes the methylation of glycine by using S-adenosylmethionine (AdoMet) to form N-methylglycine (sarcosine) with the concomitant production of S-adenosylhomocysteine (AdoHcy). Possible crucial role in the regulation of tissue concentration of AdoMet and of metabolism of methionine.
- Gene Name:
- GNMT
- Uniprot ID:
- Q14749
- Molecular Weight:
- 32742.0 Da
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Histamine n-methyltransferase activity
- Specific Function:
- Inactivates histamine by N-methylation. Plays an important role in degrading histamine and in regulating the airway response to histamine.
- Gene Name:
- HNMT
- Uniprot ID:
- P50135
- Molecular Weight:
- 33294.765 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 18.1 uM | Not Available | BindingDB 50009672 |
References
- Houston DM, Matuszewska B, Borchardt RT: Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 10. Base- and amino acid modified analogues of S-aristeromycinyl-L-homocysteine. J Med Chem. 1985 Apr;28(4):478-82. [3981540 ]
- General Function:
- Transcription regulatory region dna binding
- Specific Function:
- Methylates (mono- and asymmetric dimethylation) the guanidino nitrogens of arginyl residues in several proteins involved in DNA packaging, transcription regulation, pre-mRNA splicing, and mRNA stability. Recruited to promoters upon gene activation together with histone acetyltransferases from EP300/P300 and p160 families, methylates histone H3 at 'Arg-17' (H3R17me), forming mainly asymmetric dimethylarginine (H3R17me2a), leading to activate transcription via chromatin remodeling. During nuclear hormone receptor activation and TCF7L2/TCF4 activation, acts synergically with EP300/P300 and either one of the p160 histone acetyltransferases NCOA1/SRC1, NCOA2/GRIP1 and NCOA3/ACTR or CTNNB1/beta-catenin to activate transcription. During myogenic transcriptional activation, acts together with NCOA3/ACTR as a coactivator for MEF2C. During monocyte inflammatory stimulation, acts together with EP300/P300 as a coactivator for NF-kappa-B. Acts as coactivator for PPARG, promotes adipocyte differentiation and the accumulation of brown fat tissue. Plays a role in the regulation of pre-mRNA alternative splicing by methylation of splicing factors. Also seems to be involved in p53/TP53 transcriptional activation. Methylates EP300/P300, both at 'Arg-2142', which may loosen its interaction with NCOA2/GRIP1, and at 'Arg-580' and 'Arg-604' in the KIX domain, which impairs its interaction with CREB and inhibits CREB-dependent transcriptional activation. Also methylates arginine residues in RNA-binding proteins PABPC1, ELAVL1 and ELAV4, which may affect their mRNA-stabilizing properties and the half-life of their target mRNAs.
- Gene Name:
- CARM1
- Uniprot ID:
- Q86X55
- Molecular Weight:
- 65853.185 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.86 uM | Not Available | BindingDB 50009672 |
References
- Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y: Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J Med Chem. 2012 Sep 27;55(18):8066-74. Epub 2012 Sep 6. [22924785 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Histone methyltransferase that specifically trimethylates 'Lys-9' of histone H3 using monomethylated H3 'Lys-9' as substrate. Also weakly methylates histone H1 (in vitro). H3 'Lys-9' trimethylation represents a specific tag for epigenetic transcriptional repression by recruiting HP1 (CBX1, CBX3 and/or CBX5) proteins to methylated histones. Mainly functions in heterochromatin regions, thereby playing a central role in the establishment of constitutive heterochromatin at pericentric and telomere regions. H3 'Lys-9' trimethylation is also required to direct DNA methylation at pericentric repeats. SUV39H1 is targeted to histone H3 via its interaction with RB1 and is involved in many processes, such as repression of MYOD1-stimulated differentiation, regulation of the control switch for exiting the cell cycle and entering differentiation, repression by the PML-RARA fusion protein, BMP-induced repression, repression of switch recombination to IgA and regulation of telomere length. Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. The eNoSC complex is able to sense the energy status of cell: upon glucose starvation, elevation of NAD(+)/NADP(+) ratio activates SIRT1, leading to histone H3 deacetylation followed by dimethylation of H3 at 'Lys-9' (H3K9me2) by SUV39H1 and the formation of silent chromatin in the rDNA locus. Recruited by the large PER complex to the E-box elements of the circadian target genes such as PER2 itself or PER1, contributes to the conversion of local chromatin to a heterochromatin-like repressive state through H3 'Lys-9' trimethylation.
- Gene Name:
- SUV39H1
- Uniprot ID:
- O43463
- Molecular Weight:
- 47907.065 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 4.9 uM | Not Available | BindingDB 50009672 |
References
- Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y: Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J Med Chem. 2012 Sep 27;55(18):8066-74. Epub 2012 Sep 6. [22924785 ]
- General Function:
- Thioether s-methyltransferase activity
- Specific Function:
- Functions as thioether S-methyltransferase and is active with a variety of thioethers and the corresponding selenium and tellurium compounds, including 3-methylthiopropionaldehyde, dimethyl selenide, dimethyl telluride, 2-methylthioethylamine, 2-methylthioethanol, methyl-n-propyl sulfide and diethyl sulfide. Plays an important role in the detoxification of selenium compounds (By similarity). Catalyzes the N-methylation of tryptamine and structurally related compounds.
- Gene Name:
- INMT
- Uniprot ID:
- O95050
- Molecular Weight:
- 28890.75 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 2 uM | Not Available | BindingDB 50009672 |
References
- Benghiat E, Crooks PA: Multisubstrate adducts as potential inhibitors of S-adenosylmethionine dependent methylases: inhibition of indole N-methyltransferase by (5'-deoxyadenosyl)[3-(3-indolyl)prop-1-yl]methylsulfonium and (5'-deoxyadenosyl)[4-(3-indolyl)but-1-yl]methylsulfonium salts. J Med Chem. 1983 Oct;26(10):1470-7. [6620306 ]
- General Function:
- Protein-arginine omega-n asymmetric methyltransferase activity
- Specific Function:
- Methylates (mono and asymmetric dimethylation) the guanidino nitrogens of arginyl residues in some proteins.
- Gene Name:
- PRMT3
- Uniprot ID:
- O60678
- Molecular Weight:
- 59875.63 Da
References
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. [10592235 ]
- General Function:
- Protein-lysine n-methyltransferase activity
- Specific Function:
- Histone methyltransferase that specifically monomethylates 'Lys-4' of histone H3. H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. Plays a central role in the transcriptional activation of genes such as collagenase or insulin. Recruited by IPF1/PDX-1 to the insulin promoter, leading to activate transcription. Has also methyltransferase activity toward non-histone proteins such as p53/TP53, TAF10, and possibly TAF7 by recognizing and binding the [KR]-[STA]-K in substrate proteins. Monomethylates 'Lys-189' of TAF10, leading to increase the affinity of TAF10 for RNA polymerase II. Monomethylates 'Lys-372' of p53/TP53, stabilizing p53/TP53 and increasing p53/TP53-mediated transcriptional activation.
- Gene Name:
- SETD7
- Uniprot ID:
- Q8WTS6
- Molecular Weight:
- 40720.595 Da
- General Function:
- Trna methyltransferase activity
- Specific Function:
- Specifically methylates cytosine 38 in the anticodon loop of tRNA(Asp).
- Gene Name:
- TRDMT1
- Uniprot ID:
- O14717
- Molecular Weight:
- 44596.17 Da