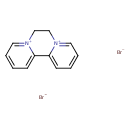Diquat dibromide (T3D4498)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2014-08-29 06:51:42 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:51 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D4498 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Diquat dibromide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Diquat dibromide is a non-selective contact herbicide, algicide, desiccant, and defoliant that produces desiccation and defoliation most often available as the dibromide, diquat dibromide. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C12H12Br2N2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 344.045 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 341.937 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 85-00-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 7λ⁵,10λ⁵-diazatricyclo[8.4.0.0²,⁷]tetradeca-1(14),2,4,6,10,12-hexaene-7,10-bis(ylium) dibromide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | diquat dication dibromide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [Br-].[Br-].C1C[N+]2=CC=CC=C2C2=CC=CC=[N+]12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C12H12N2.2BrH/c1-3-7-13-9-10-14-8-4-2-6-12(14)11(13)5-1;;/h1-8H,9-10H2;2*1H/q+2;;/p-2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=ODPOAESBSUKMHD-UHFFFAOYSA-L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pyridinium derivatives. Pyridinium derivatives are compounds containing a pyridinium ring, which is the cationic form of pyridine. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyridines and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyridinium derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyridinium derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | EYES: irrigate opened eyes for several minutes under running water. INGESTION: do not induce vomiting. Rinse mouth with water (never give anything by mouth to an unconscious person). Seek immediate medical advice. SKIN: should be treated immediately by rinsing the affected parts in cold running water for at least 15 minutes, followed by thorough washing with soap and water. If necessary, the person should shower and change contaminated clothing and shoes, and then must seek medical attention. INHALATION: supply fresh air. If required provide artificial respiration. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 6794 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL1599022 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 6536 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D4498.pdf | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Voltage-gated chloride channel activity
- Specific Function:
- Voltage-gated chloride channel. Chloride channels have several functions including the regulation of cell volume; membrane potential stabilization, signal transduction and transepithelial transport.
- Gene Name:
- CLCN1
- Uniprot ID:
- P35523
- Molecular Weight:
- 108625.435 Da
References
- Simchowitz L: Interactions of bromide, iodide, and fluoride with the pathways of chloride transport and diffusion in human neutrophils. J Gen Physiol. 1988 Jun;91(6):835-60. [3047312 ]
- Pusch M, Jordt SE, Stein V, Jentsch TJ: Chloride dependence of hyperpolarization-activated chloride channel gates. J Physiol. 1999 Mar 1;515 ( Pt 2):341-53. [10050002 ]
- General Function:
- Voltage-gated chloride channel activity
- Specific Function:
- Voltage-gated chloride channel. Chloride channels have several functions including the regulation of cell volume; membrane potential stabilization, signal transduction and transepithelial transport. May be important in urinary concentrating mechanisms.
- Gene Name:
- CLCNKA
- Uniprot ID:
- P51800
- Molecular Weight:
- 75284.08 Da
References
- Simchowitz L: Interactions of bromide, iodide, and fluoride with the pathways of chloride transport and diffusion in human neutrophils. J Gen Physiol. 1988 Jun;91(6):835-60. [3047312 ]
- Pusch M, Jordt SE, Stein V, Jentsch TJ: Chloride dependence of hyperpolarization-activated chloride channel gates. J Physiol. 1999 Mar 1;515 ( Pt 2):341-53. [10050002 ]
- General Function:
- Voltage-gated chloride channel activity
- Specific Function:
- Voltage-gated chloride channel. Chloride channels have several functions including the regulation of cell volume; membrane potential stabilization, signal transduction and transepithelial transport. May be important in urinary concentrating mechanisms.
- Gene Name:
- CLCNKB
- Uniprot ID:
- P51801
- Molecular Weight:
- 75445.3 Da
References
- Simchowitz L: Interactions of bromide, iodide, and fluoride with the pathways of chloride transport and diffusion in human neutrophils. J Gen Physiol. 1988 Jun;91(6):835-60. [3047312 ]
- Pusch M, Jordt SE, Stein V, Jentsch TJ: Chloride dependence of hyperpolarization-activated chloride channel gates. J Physiol. 1999 Mar 1;515 ( Pt 2):341-53. [10050002 ]
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel (By similarity).
- Gene Name:
- GABRA1
- Uniprot ID:
- P14867
- Molecular Weight:
- 51801.395 Da
References
- Suzuki S, Kawakami K, Nakamura F, Nishimura S, Yagi K, Seino M: Bromide, in the therapeutic concentration, enhances GABA-activated currents in cultured neurons of rat cerebral cortex. Epilepsy Res. 1994 Oct;19(2):89-97. [7843172 ]
- General Function:
- Gaba-gated chloride ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel.
- Gene Name:
- GABRB3
- Uniprot ID:
- P28472
- Molecular Weight:
- 54115.04 Da
References
- Suzuki S, Kawakami K, Nakamura F, Nishimura S, Yagi K, Seino M: Bromide, in the therapeutic concentration, enhances GABA-activated currents in cultured neurons of rat cerebral cortex. Epilepsy Res. 1994 Oct;19(2):89-97. [7843172 ]
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel.
- Gene Name:
- GABRG2
- Uniprot ID:
- P18507
- Molecular Weight:
- 54161.78 Da
References
- Suzuki S, Kawakami K, Nakamura F, Nishimura S, Yagi K, Seino M: Bromide, in the therapeutic concentration, enhances GABA-activated currents in cultured neurons of rat cerebral cortex. Epilepsy Res. 1994 Oct;19(2):89-97. [7843172 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- CCL2
- Uniprot ID:
- P13500
- Molecular Weight:
- 11024.87 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SM3C_MCP1_down | BioSeek |
| AC50 | 1.48 uM | BSK_KF3CT_MCP1_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor binding
- Specific Function:
- Chemotactic for monocytes and T-lymphocytes. Binds to CXCR3.
- Gene Name:
- CXCL10
- Uniprot ID:
- P02778
- Molecular Weight:
- 10880.915 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_KF3CT_IP10_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Cytokine activity
- Specific Function:
- Cytokine that affects the growth, movement, or activation state of cells that participate in immune and inflammatory response. Chemotactic for activated T-cells. Binds to CXCR3.
- Gene Name:
- CXCL9
- Uniprot ID:
- Q07325
- Molecular Weight:
- 14018.72 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_MIG_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Platelet-derived growth factor binding
- Specific Function:
- Collagen type III occurs in most soft connective tissues along with type I collagen. Involved in regulation of cortical development. Is the major ligand of GPR56 in the developing brain and binding to GPR56 inhibits neuronal migration and activates the RhoA pathway by coupling GPR56 to GNA13 and possibly GNA12.
- Gene Name:
- COL3A1
- Uniprot ID:
- P02461
- Molecular Weight:
- 138564.005 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_CollagenIII_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Cleaves collagens of types I, II, and III at one site in the helical domain. Also cleaves collagens of types VII and X. In case of HIV infection, interacts and cleaves the secreted viral Tat protein, leading to a decrease in neuronal Tat's mediated neurotoxicity.
- Gene Name:
- MMP1
- Uniprot ID:
- P03956
- Molecular Weight:
- 54006.61 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_BE3C_MMP1_up | BioSeek |
| AC50 | 1.48 uM | BSK_hDFCGF_MMP1_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serine-type endopeptidase inhibitor activity
- Specific Function:
- Serine protease inhibitor. This inhibitor acts as 'bait' for tissue plasminogen activator, urokinase, protein C and matriptase-3/TMPRSS7. Its rapid interaction with PLAT may function as a major control point in the regulation of fibrinolysis.
- Gene Name:
- SERPINE1
- Uniprot ID:
- P05121
- Molecular Weight:
- 45059.695 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_PAI1_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serine-type endopeptidase activity
- Specific Function:
- Converts the abundant, but inactive, zymogen plasminogen to plasmin by hydrolyzing a single Arg-Val bond in plasminogen. By controlling plasmin-mediated proteolysis, it plays an important role in tissue remodeling and degradation, in cell migration and many other physiopathological events. Plays a direct role in facilitating neuronal migration.
- Gene Name:
- PLAT
- Uniprot ID:
- P00750
- Molecular Weight:
- 62916.495 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_BE3C_tPA_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Receptor for TNFSF5/CD40LG. Transduces TRAF6- and MAP3K8-mediated signals that activate ERK in macrophages and B cells, leading to induction of immunoglobulin secretion.
- Gene Name:
- CD40
- Uniprot ID:
- P25942
- Molecular Weight:
- 30618.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SAg_CD40_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Urokinase plasminogen activator receptor activity
- Specific Function:
- Acts as a receptor for urokinase plasminogen activator. Plays a role in localizing and promoting plasmin formation. Mediates the proteolysis-independent signal transduction activation effects of U-PA. It is subject to negative-feedback regulation by U-PA which cleaves it into an inactive form.
- Gene Name:
- PLAUR
- Uniprot ID:
- Q03405
- Molecular Weight:
- 36977.62 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_BE3C_uPAR_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Involved in lymphocyte proliferation and functions as a signal transmitting receptor in lymphocytes, natural killer (NK) cells, and platelets.
- Gene Name:
- CD69
- Uniprot ID:
- Q07108
- Molecular Weight:
- 22559.25 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_CD69_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane receptor protein tyrosine phosphatase activity
- Specific Function:
- Protein tyrosine-protein phosphatase required for T-cell activation through the antigen receptor. Acts as a positive regulator of T-cell coactivation upon binding to DPP4. The first PTPase domain has enzymatic activity, while the second one seems to affect the substrate specificity of the first one. Upon T-cell activation, recruits and dephosphorylates SKAP1 and FYN. Dephosphorylates LYN, and thereby modulates LYN activity (By similarity).
- Gene Name:
- PTPRC
- Uniprot ID:
- P08575
- Molecular Weight:
- 147253.075 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.40 uM | NVS_ENZ_hPTPRC | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]