| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-08-29 06:51:44 UTC |
|---|
| Update Date | 2014-12-24 20:26:52 UTC |
|---|
| Accession Number | T3D4519 |
|---|
| Identification |
|---|
| Common Name | Malaoxon |
|---|
| Class | Small Molecule |
|---|
| Description | Malaoxon is a metabolite of malathion. Malaoxon is a chemical compound with the formula C10H19O7PS. More specifically, it is a phosphorothioate. It is a breakdown product of, and more toxic than, malathion. |
|---|
| Compound Type | - Animal Toxin
- Ester
- Ether
- Metabolite
- Natural Compound
- Organic Compound
|
|---|
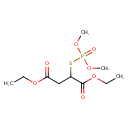
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 2-[(Dimethoxyphosphinyl)thio]butanedioic acid | | Carbethoxy malaoxon | | Diethyl 2-[(dimethoxyphosphoryl)sulfanyl]succinate | | Diethyl 2-[(dimethoxyphosphoryl)thio]succinate | | Diethyl((dimethoxyphosphinyl)thio)butanedioate | | liromat | | Malaoxone | | O,O-Dimethyl S-(1,2-dicarbethoxy)ethyl phosphorothioate | | O,O-dimethyl S-1,2-bis(ethoxycarbonyl)ethyl phosphorothioate | | O,O-dimethyl-S-(1,2-dicarbethoxy)ethyl phosphorothioate |
|
|---|
| Chemical Formula | C10H19O7PS |
|---|
| Average Molecular Mass | 314.292 g/mol |
|---|
| Monoisotopic Mass | 314.059 g/mol |
|---|
| CAS Registry Number | 1634-78-2 |
|---|
| IUPAC Name | 1,4-diethyl 2-[(dimethoxyphosphoryl)sulfanyl]butanedioate |
|---|
| Traditional Name | malaoxon |
|---|
| SMILES | CCOC(=O)CC(SP(=O)(OC)OC)C(=O)OCC |
|---|
| InChI Identifier | InChI=1/C10H19O7PS/c1-5-16-9(11)7-8(10(12)17-6-2)19-18(13,14-3)15-4/h8H,5-7H2,1-4H3 |
|---|
| InChI Key | InChIKey=WSORODGWGUUOBO-UHFFFAOYNA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as fatty acid esters. These are carboxylic ester derivatives of a fatty acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acid esters |
|---|
| Direct Parent | Fatty acid esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Fatty acid ester
- Dicarboxylic acid or derivatives
- Carboxylic acid ester
- Sulfenyl compound
- Organothiophosphorus compound
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organosulfur compound
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-4490000000-3be32b99d94a0568dba5 | 2017-09-20 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-004i-1900000000-f245f9049a6ed189a87f | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-002b-9800000000-bc654e513565551ef2f0 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0002-9200000000-5f8da96f7a4bd6c28f2d | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0002-9100000000-92717e126efe9cd1dbe9 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0002-9100000000-3fa546574076e3dc68f3 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-006t-9100000000-bbf30244ec610d5312e7 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-00dj-9000000000-4d8154797f79dfbdfd82 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0fk9-9000000000-713f99c243580257491a | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , positive | splash10-0udi-9000000000-407ae6fac33cef57b3ca | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 90V, Positive | splash10-006t-9100000000-13433d2b98aac0799930 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 60V, Positive | splash10-0002-9100000000-1571d4d0edb12dfcb446 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 75V, Positive | splash10-0002-9100000000-01d68a9fcce063a364db | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 15V, Positive | splash10-004i-1900000000-e66047fab58bcb4d3f0b | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 45V, Positive | splash10-0002-9200000000-fd95bdd28ee3debbbd2f | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-002b-9800000000-0a3e8dfdfb86454ce2a8 | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0192000000-245618c3e5fc21daa081 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9580000000-728ce87c907b04b594b0 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-1930000000-eb1eb9fd2804f9750fe4 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-02t9-1192000000-34cd3addfa580ad01b8a | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-07p4-0190000000-f405e4606435882da7cf | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-114j-1290000000-bb2beb159ce8bc944699 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00b9-0901000000-6c82ff5aadf81a58e0d1 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-016r-0920000000-88556df83a495bae7d7f | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08ml-4900000000-d4c35b02d04ed8fc2af2 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0900000000-870ef291d60f89042637 | 2021-10-12 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-24 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Malaoxon is a cholinesterase or acetylcholinesterase (AChE) inhibitor. A cholinesterase inhibitor (or 'anticholinesterase') suppresses the action of acetylcholinesterase. Because of its essential function, chemicals that interfere with the action of acetylcholinesterase are potent neurotoxins, causing excessive salivation and eye-watering in low doses, followed by muscle spasms and ultimately death. Nerve gases and many substances used in insecticides have been shown to act by binding a serine in the active site of acetylcholine esterase, inhibiting the enzyme completely. Acetylcholine esterase breaks down the neurotransmitter acetylcholine, which is released at nerve and muscle junctions, in order to allow the muscle or organ to relax. The result of acetylcholine esterase inhibition is that acetylcholine builds up and continues to act so that any nerve impulses are continually transmitted and muscle contractions do not stop. Among the most common acetylcholinesterase inhibitors are phosphorus-based compounds, which are designed to bind to the active site of the enzyme. The structural requirements are a phosphorus atom bearing two lipophilic groups, a leaving group (such as a halide or thiocyanate), and a terminal oxygen. |
|---|
| Metabolism | Metabolism of organophosphates occurs principally by oxidation, by hydrolysis via esterases and by reaction with glutathione. Demethylation and glucuronidation may also occur. Oxidation of organophosphorus pesticides may result in moderately toxic products. In general, phosphorothioates are not directly toxic but require oxidative metabolism to the proximal toxin. The glutathione transferase reactions produce products that are, in most cases, of low toxicity. Paraoxonase (PON1) is a key enzyme in the metabolism of organophosphates. PON1 can inactivate some organophosphates through hydrolysis. PON1 hydrolyzes the active metabolites in several organophosphates insecticides as well as, nerve agents such as soman, sarin, and VX. The presence of PON1 polymorphisms causes there to be different enzyme levels and catalytic efficiency of this esterase, which in turn suggests that different individuals may be more susceptible to the toxic effect of organophosphate exposure. |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Acute exposure to cholinesterase inhibitors can cause a cholinergic crisis characterized by severe nausea/vomiting, salivation, sweating, bradycardia, hypotension, collapse, and convulsions. Increasing muscle weakness is a possibility and may result in death if respiratory muscles are involved. Accumulation of ACh at motor nerves causes overstimulation of nicotinic expression at the neuromuscular junction. When this occurs symptoms such as muscle weakness, fatigue, muscle cramps, fasciculation, and paralysis can be seen. When there is an accumulation of ACh at autonomic ganglia this causes overstimulation of nicotinic expression in the sympathetic system. Symptoms associated with this are hypertension, and hypoglycemia. Overstimulation of nicotinic acetylcholine receptors in the central nervous system, due to accumulation of ACh, results in anxiety, headache, convulsions, ataxia, depression of respiration and circulation, tremor, general weakness, and potentially coma. When there is expression of muscarinic overstimulation due to excess acetylcholine at muscarinic acetylcholine receptors symptoms of visual disturbances, tightness in chest, wheezing due to bronchoconstriction, increased bronchial secretions, increased salivation, lacrimation, sweating, peristalsis, and urination can occur. Certain reproductive effects in fertility, growth, and development for males and females have been linked specifically to organophosphate pesticide exposure. Most of the research on reproductive effects has been conducted on farmers working with pesticides and insecticdes in rural areas. In females menstrual cycle disturbances, longer pregnancies, spontaneous abortions, stillbirths, and some developmental effects in offspring have been linked to organophosphate pesticide exposure. Prenatal exposure has been linked to impaired fetal growth and development. Neurotoxic effects have also been linked to poisoning with OP pesticides causing four neurotoxic effects in humans: cholinergic syndrome, intermediate syndrome, organophosphate-induced delayed polyneuropathy (OPIDP), and chronic organophosphate-induced neuropsychiatric disorder (COPIND). These syndromes result after acute and chronic exposure to OP pesticides. |
|---|
| Symptoms | Symptoms of low dose exposure include excessive salivation and eye-watering. Acute dose symptoms include severe nausea/vomiting, salivation, sweating, bradycardia, hypotension, collapse, and convulsions. Increasing muscle weakness is a possibility and may result in death if respiratory muscles are involved. Hypertension, hypoglycemia, anxiety, headache, tremor and ataxia may also result. |
|---|
| Treatment | If the compound has been ingested, rapid gastric lavage should be performed using 5% sodium bicarbonate. For skin contact, the skin should be washed with soap and water. If the compound has entered the eyes, they should be washed with large quantities of isotonic saline or water. In serious cases, atropine and/or pralidoxime should be administered. Anti-cholinergic drugs work to counteract the effects of excess acetylcholine and reactivate AChE. Atropine can be used as an antidote in conjunction with pralidoxime or other pyridinium oximes (such as trimedoxime or obidoxime), though the use of '-oximes' has been found to be of no benefit, or possibly harmful, in at least two meta-analyses. Atropine is a muscarinic antagonist, and thus blocks the action of acetylcholine peripherally. |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB60627 |
|---|
| PubChem Compound ID | Not Available |
|---|
| ChEMBL ID | CHEMBL1331476 |
|---|
| ChemSpider ID | Not Available |
|---|
| KEGG ID | Not Available |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | Not Available |
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|