| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-09-11 05:19:19 UTC |
|---|
| Update Date | 2014-12-24 20:26:58 UTC |
|---|
| Accession Number | T3D4853 |
|---|
| Identification |
|---|
| Common Name | 3-Methyl-4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-3-buten-2-one |
|---|
| Class | Small Molecule |
|---|
| Description | (±)-(E)-3-Methyl-4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-3-buten-2-one is a flavouring ingredient
3-Methyl-4-(2,6,6-trimethyl-2-cyclohexen-1-yl)-3-buten-2-one belongs to the family of Monoterpenes. These are compounds contaning a chain of two isoprene units. |
|---|
| Compound Type | - Ester
- Flavouring Agent
- Food Toxin
- Household Toxin
- Metabolite
- Organic Compound
- Synthetic Compound
|
|---|
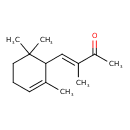
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 3-Methyl-a-ionone | | Cetone alpha | | FEMA 2714 | | Isomethyl-a-ionone |
|
|---|
| Chemical Formula | C14H22O |
|---|
| Average Molecular Mass | 206.324 g/mol |
|---|
| Monoisotopic Mass | 206.167 g/mol |
|---|
| CAS Registry Number | 127-51-5 |
|---|
| IUPAC Name | (3E)-3-methyl-4-(2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-one |
|---|
| Traditional Name | (3E)-3-methyl-4-(2,6,6-trimethylcyclohex-2-en-1-yl)but-3-en-2-one |
|---|
| SMILES | [H]\C(C1C(C)=CCCC1(C)C)=C(\C)C(C)=O |
|---|
| InChI Identifier | InChI=1/C14H22O/c1-10-7-6-8-14(4,5)13(10)9-11(2)12(3)15/h7,9,13H,6,8H2,1-5H3/b11-9+ |
|---|
| InChI Key | InChIKey=JRJBVWJSTHECJK-PKNBQFBNNA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as sesquiterpenoids. These are terpenes with three consecutive isoprene units. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Sesquiterpenoids |
|---|
| Direct Parent | Sesquiterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cyclofarsesane sesquiterpenoid
- Megastigmane sesquiterpenoid
- Sesquiterpenoid
- Ionone derivative
- Alpha-branched alpha,beta-unsaturated-ketone
- Alpha,beta-unsaturated ketone
- Enone
- Acryloyl-group
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01ox-4900000000-188a75ba35cc10a457d4 | 2016-09-22 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-3980000000-aecfe987633b1b3253c9 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05n0-5910000000-d30c3fd0fb817411950b | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uxr-9100000000-9b4de8add50fbadeab2b | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0190000000-832ea298f97bab583ddc | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bt9-3890000000-f8b94bab666cc1f09e7c | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-02ga-4900000000-aab4a14210d641ae936f | 2016-08-04 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0490000000-b8904ddd6a4476e42b5f | 2021-09-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fr-0930000000-c948347582e5019af52e | 2021-09-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00l6-9100000000-02cc3f9860c78776dde2 | 2021-09-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1910000000-96bf5af4c798ec733d36 | 2021-09-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00xr-7900000000-f65ac4d5c7d66a0b1bad | 2021-09-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05mo-9300000000-1f95634a65fdfd95d3c4 | 2021-09-22 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Not Available |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB31738 |
|---|
| PubChem Compound ID | 5372174 |
|---|
| ChEMBL ID | Not Available |
|---|
| ChemSpider ID | 4522510 |
|---|
| KEGG ID | Not Available |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | - Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC.
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|