| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-06-01 19:33:01 UTC |
|---|
| Update Date | 2014-12-24 20:22:50 UTC |
|---|
| Accession Number | T3D0797 |
|---|
| Identification |
|---|
| Common Name | Imidacloprid |
|---|
| Class | Small Molecule |
|---|
| Description | Imidacloprid is a neonicotinoid insecticide, which is a class of neuro-active insecticides modeled after nicotine. Nicotine was identified and used as an insecticide and rat poison as early as the 1600’s. Its effectiveness as an insecticide spurred a search for insecticidal compounds that have selectively less effect on mammals, which led to the discovery of neonicotinoids. Neonicotinoids, like nicotine, bind to nicotinic acetylcholine receptors of a cell. In mammals, nicotinic acetylcholine receptors are located in cells of both the central and peripheral nervous systems. In insects these receptors are limited to the CNS. While low to moderate activation of these receptors causes nervous stimulation, high levels overstimulate and block the receptors causing paralysis and death. Nicotinic acetylcholine receptors are activated by the neurotransmitter acetylcholine. Acetylcholine is broken down by acetylcholinesterase to terminate signals from these receptors. However, acetylcholinesterase cannot break down neonicotinoids and the binding is irreversible. Because most neonicotinoids bind much more strongly to insect neuron receptors than to mammal neuron receptors, these insecticides are selectively more toxic to insects than mammals. The low mammalian toxicity of neonicotinoids can be explained in large part by their lack of a charged nitrogen atom at physiological pH. The uncharged molecule can penetrate the insect blood–brain barrier, while the mammalian blood–brain barrier filters it. However, Some neonicotinoid breakdown products are toxic to humans, especially if they have become charged. Because of their low toxicity and other favorable features, neonicotinoids are among the most widely used insecticides in the world. Most neonicotinoids are water-soluble and break down slowly in the environment, so they can be taken up by the plant and provide protection from insects as the plant grows. Neonicotinoids are currently used on corn, canola, cotton, sorghum, sugar beets and soybeans. They are also used on the vast majority of fruit and vegetable crops, including apples, cherries, peaches, oranges, berries, leafy greens, tomatoes, and potatoes. The use of neonicotinoids has been linked in a range of studies to adverse ecological effects, including honey-bee colony collapse disorder (CCD) and loss of birds due to a reduction in insect populations. This has led to moratoriums and bans on their use in Europe. |
|---|
| Compound Type | - Amide
- Amine
- Aromatic Hydrocarbon
- Food Toxin
- Household Toxin
- Insecticide
- Metabolite
- Neonicotinoid
- Organic Compound
- Organochloride
- Pesticide
- Synthetic Compound
|
|---|
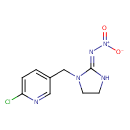
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (e)-Imidacloprid | | (z)-Imidacloprid | | 1-((6-Chloro-3-pyridyl)methyl)-N-nitro-2-imidazolidinimine | | 1-(2-Chloro-5-pyridylmethyl)-2-(nitroimino)imidazolidine | | 1-[(6-Chloro-3-pyridinyl)methyl]-4,5-dihydro-N-nitro-1H-imidazol-2-amine | | Admire | | Advantage | | Confidor | | Confidor 200 SL | | Confidor SL | | Gaucho | | Imazethapyr | | Imidacloprid (old RN) | | Merit | | Premise 75 | | Provado |
|
|---|
| Chemical Formula | C9H10ClN5O2 |
|---|
| Average Molecular Mass | 255.661 g/mol |
|---|
| Monoisotopic Mass | 255.052 g/mol |
|---|
| CAS Registry Number | 105827-78-9 |
|---|
| IUPAC Name | 2-chloro-5-{[2-(nitroamino)-4,5-dihydro-1H-imidazol-1-yl]methyl}pyridine |
|---|
| Traditional Name | imidacloprid |
|---|
| SMILES | ClC1=NC=C(CN2CCN=C2NN(=O)=O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C9H10ClN5O2/c10-8-2-1-7(5-12-8)6-14-4-3-11-9(14)13-15(16)17/h1-2,5H,3-4,6H2,(H,11,13) |
|---|
| InChI Key | InChIKey=YWTYJOPNNQFBPC-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as nitroguanidines. These are organonitrogen compounds containing a nitro group, which is N-linked to a guanidine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Guanidines |
|---|
| Direct Parent | Nitroguanidines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Nitroguanidine
- 2-halopyridine
- Aryl chloride
- Aryl halide
- Pyridine
- 2-imidazoline
- Nitramine
- Heteroaromatic compound
- Organic nitro compound
- Azacycle
- Carboximidamide
- Allyl-type 1,3-dipolar organic compound
- Propargyl-type 1,3-dipolar organic compound
- Organic 1,3-dipolar compound
- Organoheterocyclic compound
- Organochloride
- Organic oxygen compound
- Organic zwitterion
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organohalogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | Colorless crystals (5). |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 143 - 144°C | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004i-2910000000-16207f958c3941d24901 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-05di-2920000000-c29270e5a1a153c5bef7 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0udi-0290000000-0812870c582a68d58f9e | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0930000000-343db00f5be70f0fe78c | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0090000000-719eaaca2c7a38c90d68 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0090000000-942ea462298844bafdc2 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0090000000-56328a93fd1e38aa2242 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-000i-9000000000-cad2f4a13e5493517e85 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-000i-9000000000-5f6d2d344bf6659eca47 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0090000000-68418a137b14a769367c | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0090000000-47ad82be374a283a9dbb | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0udi-0920000000-46ef972fc42dceee7393 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QFT , negative | splash10-0udi-0090000000-fc3fa48055c83ff1de5f | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a4i-0190000000-0cd6e54fe940f97ccb1d | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-056r-0980000000-e48625fbd18b2d626cc6 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-056r-0960000000-89d9de4658faefa962b1 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a6r-0940000000-16fe6279a6031f08bb67 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0aba-0910000000-2859fa22e79d10470b94 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-08fr-0390000000-ab06f369da2af69be924 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0a4i-0190000000-af26c2abec118b42cc59 | 2017-09-14 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0090000000-3250c0b819f9c6714e58 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4j-0090000000-e8976c6ed0aab7cb6f36 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001l-7910000000-216222493c38e9ac9d30 | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-3090000000-b3146d60a6f2b5d2900c | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-2190000000-54519332ebe8fd4cbef0 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-b63d477ace752edc00a8 | 2016-08-03 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | Not Available | 2014-09-20 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100.40 MHz, CDCl3, experimental) | Not Available | 2014-09-23 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Inhalation (5) ; dermal (5) ; oral (5) |
|---|
| Mechanism of Toxicity | Imidacloprid acts on the nicotinic acetylcholine receptor; the chlorination inhibits degradation by acetylcholine-esterase (5). |
|---|
| Metabolism | Two main routes of metabolism responsible for the degradation of imidacloprid were identified. The first is oxidative cleavage, yielding 6-chloronicotinic acid, which is conjugated with glycine to form a hippuric acid-type conjugate. These two metabolites together represented most of the identified metabolites, or about 30% of the recovered radiolabel. Of minor importance in terms of quantity is dechlorination of the pyridinyl moiety, producing the 6-hydroxy nicotinic acid and its methylmercapturic acid derivative, probably as a degradation product of a glutathione conjugate. The 6-methylmercapto nicotinic acid conjugated with glycine, and the glycine conjugate constituted 5.6% of the recovered radiolabel. The second important biodegradation step starts with hydroxylation of the imidazolidine ring at the 4 or 5 position, and about 16% of the recovered radiolabel was identified as the sum of 4- and 5-hydroxy imidacloprid. The loss of water yields the olefinic compound. These biotransformation products and the unchanged parent compound were excreted in urine and feces, while the guanidine compound was a less important metabolite and was eliminated only in feces (1). |
|---|
| Toxicity Values | LD50: 450 mg/kg (Oral, Rat) (5)
LD50: 131 mg/kg (Oral, Mouse) (5)
LD50: >5000 mg/kg (Dermal, Rat) (5)
LD50: 69 mg/m3 (Inhalation (aerosol), Rat) (5)
LD50: 5 323 mg/m3 (Inhalation (dust), Rat) (5) |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | The most widely used applications for Imidacloprid in California are pest control in structures, turf pest control, grape growing, and head and leaf lettuce growing. Other widespread crop uses are rice, grains/cereals (5). |
|---|
| Minimum Risk Level | Chronic Oral: 72 mg/kg/day (Rabbit) (5) |
|---|
| Health Effects | Hypotension, fatal ventricular dysrhythmias, tremors, impaired pupillary function, and hypothermia may result from Imidacloprid poisoning (2). |
|---|
| Symptoms | Fatigue, twitching, cramps, and weakness leading to asphyxia (5). |
|---|
| Treatment | There is no known antidote for neonicotinoid insecticide exposures. The treatment is supportive and symptomatic. Administer charcoal as a slurry. In case of inhalation, move patient to fresh air, monitor for respiratory distress. If the exposure occurred via eye contact, irrigate exposed eyes with copious amounts of room temperature water for at least 15 minutes. Remove contaminated clothing and wash exposed area thoroughly with soap and water if the exposure occurred via dermal contact. (2)
|
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB40292 |
|---|
| PubChem Compound ID | 86418 |
|---|
| ChEMBL ID | Not Available |
|---|
| ChemSpider ID | 77934 |
|---|
| KEGG ID | C11110 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 5870 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Imidacloprid |
|---|
| PDB ID | IM4 |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Imidacloprid |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D0797.pdf |
|---|
| General References | - de Jongh R, Groenink L, van Der Gugten J, Olivier B: The light-enhanced startle paradigm as a putative animal model for anxiety: effects of chlordiazepoxide, flesinoxan and fluvoxamine. Psychopharmacology (Berl). 2002 Jan;159(2):176-80. Epub 2001 Sep 22. [11862346 ]
- Rumack BH (2009). POISINDEX(R) Information System. Englewood, CO: Micromedex, Inc. CCIS Volume 141, edition expires Aug, 2009.
- WHO/FAO (2001). Joint Meeting on Pesticide Residues on Imidacloprid (138261-41-3).
- Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC.
- Wikipedia. Imidacloprid. Last Updated 7 August 2009. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|