| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-06-24 17:37:26 UTC |
|---|
| Update Date | 2014-12-24 20:25:13 UTC |
|---|
| Accession Number | T3D2093 |
|---|
| Identification |
|---|
| Common Name | 1,2,3,7,8-Pentachlorodibenzo-p-dioxin |
|---|
| Class | Small Molecule |
|---|
| Description | 1,2,3,7,8-Pentachlorodibenzo-p-dioxin is a chlorinated dibenzo-p-dioxin (CDD) isomer. CDDs are a class of manufactured chemicals that consist of dioxin skeletal structures with chlorine substituents. They are also persistent organic pollutants (POPs), thus their production is regulated in most areas. Dioxins occur as by-products from the manufacture of organochlorides, the bleaching of paper, chlorination by waste and drinking water treatment plants, municipal solid waste and industrial incinerators, and natural sources such as volcanoes and forest fires. (3, 4) |
|---|
| Compound Type | - Aromatic Hydrocarbon
- Chlorinated Dibenzo-p-dioxin
- Ether
- Industrial By-product/Pollutant
- Industrial/Workplace Toxin
- Organic Compound
- Organochloride
- Pollutant
- Synthetic Compound
|
|---|
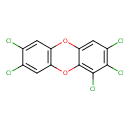
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 1,2,3,7,8-PeCDD | | 1,2,3,7,8-Penta polychlorinated dibenzo-p-dioxin | | 1,2,3,7,8-Pentachlorodibenzo(b,e)(1,4)dioxin | | 1,2,3,7,8-Pentachlorodibenzo-P-dioxin | | 1,2,3,7,8-Pentachlorooxanthrene | | PCDD 54 |
|
|---|
| Chemical Formula | C12H3Cl5O2 |
|---|
| Average Molecular Mass | 356.416 g/mol |
|---|
| Monoisotopic Mass | 353.858 g/mol |
|---|
| CAS Registry Number | 40321-76-4 |
|---|
| IUPAC Name | 1,2,3,7,8-pentachlorooxanthrene |
|---|
| Traditional Name | 1,2,3,7,8-pentachlorooxanthrene |
|---|
| SMILES | ClC1=CC2=C(OC3=C(Cl)C(Cl)=C(Cl)C=C3O2)C=C1Cl |
|---|
| InChI Identifier | InChI=1S/C12H3Cl5O2/c13-4-1-7-8(2-5(4)14)19-12-9(18-7)3-6(15)10(16)11(12)17/h1-3H |
|---|
| InChI Key | InChIKey=FSPZPQQWDODWAU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as chlorinated dibenzo-p-dioxins. These are organic compounds containing a chlorine atom attached to a dibenzo-p-dioxin moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Benzodioxins |
|---|
| Sub Class | Benzo-p-dioxins |
|---|
| Direct Parent | Chlorinated dibenzo-p-dioxins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Chlorinated-dibenzo-p-dioxin
- Diaryl ether
- Benzenoid
- Aryl halide
- Aryl chloride
- Oxacycle
- Ether
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organochloride
- Organohalogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | Colorless solid. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 240°C | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-3079000000-b5a30ae4fe361e5b341b | 2021-09-23 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0009000000-a979278bc1bad0d1a2bc | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0009000000-a979278bc1bad0d1a2bc | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0009000000-96b5ac506bd6457dfe77 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-484bbfdb53eeac513fb2 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0009000000-1fe3613b59fce8994277 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uec-6098000000-990c8a26f11d5f321cfc | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0009000000-01da1ec8e5faea7e4ef9 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0009000000-01da1ec8e5faea7e4ef9 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0009000000-01da1ec8e5faea7e4ef9 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0009000000-fa928a3b09ed103fb3d0 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0009000000-fa928a3b09ed103fb3d0 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f6x-0097000000-ec89fe3c358f5c2ae856 | 2021-10-12 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0a4i-0139000000-9561997a66bec5a473c7 | 2014-09-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Oral (3) ; inhalation(3) ; dermal (3) |
|---|
| Mechanism of Toxicity | CDDs cause their toxic effects by binding to the aryl hydrocarbon receptor and subsequently altering the transcription of certain genes. The affinity for the Ah receptor depends on the structure of the specific CDD. The change in gene expression may result from the direct interaction of the Ah receptor and its heterodimer-forming partner, the aryl hydrocarbon receptor nuclear translocator, with gene regulatory elements or the initiation of a phosphorylation/dephosphorylation cascade that subsequently activates other transcription factors. The affected genes include several oncogenes, growth factors, receptors, hormones, and drug-metabolizing enzymes. The change in transcription/translation of these genes is believed to be the cause of most of the toxic effects of CDDs. (3) |
|---|
| Metabolism | CDDs are absorbed through oral, inhalation, and dermal routes of exposure. CDDs are carried in the plasma by serum lipids and lipoproteins, distributing mainly to the liver and adipose tissue. CDDs are very slowly metabolized by the microsomal monooxygenase system to polar metabolites that can undergo conjugation with glucuronic acid and glutathione. They may increase the rate of their own metabolism by inducing both phase I and phase II enzymes. The major routes of excretion of CDDs are the bile and the faeces, though smaller amounts are excreted in the urine and via lactation. (3) |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | 3, not classifiable as to its carcinogenicity to humans. (2) |
|---|
| Uses/Sources | Dioxins occur as by-products from the manufacture of organochlorides, the bleaching of paper, chlorination by waste and drinking water treatment plants, municipal solid waste and industrial incinerators, and natural sources such as volcanoes and forest fires. (3, 4) |
|---|
| Minimum Risk Level | Acute Oral: 0.0002 ug/kg/day (1)
Intermediate Oral: 0.00002 ug/kg/day (1)
Chronic Oral: 0.000001 ug/kg/day (1) |
|---|
| Health Effects | Exposure to large amounts of CDDs causes chloracne, a severe skin disease with acne-like lesions that occur mainly on the face and upper body. CDDs may also cause liver damage and induce long-term alterations in glucose metabolism and subtle changes in hormonal levels. In addition, studies have shown that CDDs may disrupt the endocrine system and weaken the immune system, as well as cause reproductive damage and birth defects, central and peripheral nervous system pathology, thyroid disorders, endometriosis, and diabetes. (3, 4) |
|---|
| Symptoms | In addition to chloracne, CDD exposure causes skin rashes, discoloration, and excessive body hair. (3) |
|---|
| Treatment | Treatment of CDD exposure may include washing the area of contact, GI decontamination, administering an IV, or forced alkaline diuresis. (5) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 38439 |
|---|
| ChEMBL ID | CHEMBL137220 |
|---|
| ChemSpider ID | 35228 |
|---|
| KEGG ID | C18099 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | DIHYDRO-DIOH-BENZOATE |
|---|
| CTD ID | C051452 |
|---|
| Stitch ID | 1,2,3,7,8-Pentachlorodibenzo-p-dioxin |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | 7991 |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | - ATSDR - Agency for Toxic Substances and Disease Registry (2001). Minimal Risk Levels (MRLs) for Hazardous Substances. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- International Agency for Research on Cancer (2014). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Link]
- ATSDR - Agency for Toxic Substances and Disease Registry (1998). Toxicological profile for chlorinated dibenzo-p-dioxins (CDDs). U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- Wikipedia. Polychlorinated dibenzodioxins. Last Updated 19 May 2009. [Link]
- US Environmental Protection Agency (2009). Recognition and Management of Pesticide Poisonings. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | | Gene | Gene Symbol | Gene ID | Interaction | Chromosome | Details |
|---|
|
|---|
| Down-Regulated Genes | Not Available |
|---|