| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-03 22:19:14 UTC |
|---|
| Update Date | 2014-12-24 20:25:40 UTC |
|---|
| Accession Number | T3D2532 |
|---|
| Identification |
|---|
| Common Name | Arenobufagin |
|---|
| Class | Small Molecule |
|---|
| Description | Arenobufagin is a cardiotoxic bufanolide steroid secreted by the Argentine toad (Bufo arenarum). It blocks the sodium/potassium pump in heart tissue. (2) |
|---|
| Compound Type | - Animal Toxin
- Ester
- Frog/Toad Toxin
- Natural Compound
- Organic Compound
|
|---|
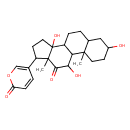
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 5-[(3S,5R,10S,11S,13R,14S,17R)-3,11,14-trihydroxy-10,13-dimethyl-12-oxo-2,3,4,5,6,7,8,9,11,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]pyran-2-one |
|
|---|
| Chemical Formula | C24H32O6 |
|---|
| Average Molecular Mass | 416.507 g/mol |
|---|
| Monoisotopic Mass | 416.220 g/mol |
|---|
| CAS Registry Number | 464-74-4 |
|---|
| IUPAC Name | 5-{5,11,17-trihydroxy-2,15-dimethyl-16-oxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl}-2H-pyran-2-one |
|---|
| Traditional Name | arenobufagin |
|---|
| SMILES | CC12C(CCC1(O)C1CCC3CC(O)CCC3(C)C1C(O)C2=O)C1=COC(=O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C24H32O6/c1-22-9-7-15(25)11-14(22)4-5-17-19(22)20(27)21(28)23(2)16(8-10-24(17,23)29)13-3-6-18(26)30-12-13/h3,6,12,14-17,19-20,25,27,29H,4-5,7-11H2,1-2H3 |
|---|
| InChI Key | InChIKey=JGDCRWYOMWSTFC-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as bufanolides and derivatives. These are steroid lactones containing a pyran-2-one moiety linked to the C17 atom of a cyclopenta[a]phenanthrene derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Steroid lactones |
|---|
| Direct Parent | Bufanolides and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Bufanolide-skeleton
- 3-hydroxysteroid
- 14-hydroxysteroid
- Hydroxysteroid
- 12-oxosteroid
- Oxosteroid
- 11-hydroxysteroid
- Pyranone
- Pyran
- Tertiary alcohol
- Heteroaromatic compound
- Cyclic alcohol
- Secondary alcohol
- Ketone
- Lactone
- Polyol
- Oxacycle
- Organoheterocyclic compound
- Alcohol
- Carbonyl group
- Organic oxide
- Hydrocarbon derivative
- Organic oxygen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000b-0914000000-6b19a919f97c568c2904 | 2021-09-24 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | 2021-11-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00l2-0009200000-ec9bea7f168e7ad8d0ea | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001j-0209000000-d99e506bdb1224c9cc5d | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-015c-2409000000-571414e2629a135d205b | 2016-08-01 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0005900000-13d84e9eb810c8cc56f7 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00kb-1009400000-e39a1d1a31283c7dd4e9 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0q2c-2009000000-851242390aab2495002a | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-0006900000-9112f39773da4209d0b0 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00l2-0109100000-74ba32d7e2de994fdb32 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-006w-3905000000-f9b793d143d1142df64d | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000900000-9a890c6be382e03ff0e6 | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0206900000-a7500363755fd25629ef | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-05ow-6595100000-0fa57da16ad9cd24787a | 2021-10-12 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Injection (sting/bite) (4) ; inhalation (smoking) (5) |
|---|
| Mechanism of Toxicity | Arenobufagin blocks the sodium/potassium pump in heart tissue. (2) |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Arenobufagin is secreted by the Argentine toad (Bufo arenarum). (2) |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Arenobufagin is cardiotoxic. (2) |
|---|
| Symptoms | Symptoms include nausea, vomiting, anorexia, diarrhoea, abdominal pain, wild hallucinations, delirium, and severe headache. Depending on the severity the victim may later suffer irregular and slow pulse, tremors, various cerebral disturbances, especially of a visual nature, convulsions, and deadly disturbances of the heart such as heart block, brachycardia, or tachycardia. (2, 3) |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 10051 |
|---|
| ChEMBL ID | Not Available |
|---|
| ChemSpider ID | Not Available |
|---|
| KEGG ID | Not Available |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | C055393 |
|---|
| Stitch ID | Arenobufagin |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | Not Available |
|---|
| General References | - Wikipedia. 2,4-Dinitrotoluene. Last Updated 10 June 2009. [Link]
- Wikipedia. Arenobufagin. Last Updated 23 January 2009. [Link]
- Wikipedia. Digitalis. Last Updated 29 June 2009. [Link]
- Wikipedia. Snake venom. Last Updated 25 July 2009. [Link]
- Wikipedia. Frog. Last Updated 10 August 2009. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|