| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-15 20:48:38 UTC |
|---|
| Update Date | 2014-12-24 20:25:49 UTC |
|---|
| Accession Number | T3D2679 |
|---|
| Identification |
|---|
| Common Name | Nitrofurantoin |
|---|
| Class | Small Molecule |
|---|
| Description | A bacteriostatic or bactericidal agent depending on the concentration and susceptibility of the infecting organism. Nitrofurantoin is active against some gram positive organisms such as S. aureus, S. epidermidis, S. saprophyticus, Enterococcus faecalis, S. agalactiae, group D streptococci, viridians streptococci and Corynebacterium. Its spectrum of activity against gram negative organisms includes E. coli, Enterobacter, Neisseria, Salmonella and Shigella. It may be used as an alternative to trimethoprim/sulfamethoxazole for treating urinary tract infections though it may be less effective at eradicating vaginal bacteria. May also be used in females as prophylaxis against recurrent cystitis related to coitus. Nitrofurantoin is highly stable to the development of bacterial resistance, a property thought to be due to its multiplicity of mechanisms of action. |
|---|
| Compound Type | - Amide
- Amine
- Anti-Infective Agent
- Anti-Infective Agent, Urinary
- Drug
- Metabolite
- Organic Compound
- Synthetic Compound
|
|---|
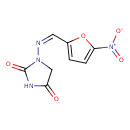
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 5-Nitrofurantoin | | Furabid | | Furadantin | | Macrobid | | Macrodantin | | N-(5-Nitrofurfurylidene)-1-aminohydantoin | | Niftran | | Siraliden | | Urantoin | | Urolong |
|
|---|
| Chemical Formula | C8H6N4O5 |
|---|
| Average Molecular Mass | 238.157 g/mol |
|---|
| Monoisotopic Mass | 238.034 g/mol |
|---|
| CAS Registry Number | 67-20-9 |
|---|
| IUPAC Name | 1-[(Z)-[(5-nitrofuran-2-yl)methylidene]amino]imidazolidine-2,4-dione |
|---|
| Traditional Name | nitrofurantoin |
|---|
| SMILES | [H]\C(=N\N1CC(O)=NC1=O)C1=CC=C(O1)N(=O)=O |
|---|
| InChI Identifier | InChI=1S/C8H6N4O5/c13-6-4-11(8(14)10-6)9-3-5-1-2-7(17-5)12(15)16/h1-3H,4H2,(H,10,13,14)/b9-3- |
|---|
| InChI Key | InChIKey=NXFQHRVNIOXGAQ-OQFOIZHKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydantoins. These are heterocyclic compounds containing an imidazolidine substituted by ketone group at positions 2 and 4. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Azolidines |
|---|
| Sub Class | Imidazolidines |
|---|
| Direct Parent | Hydantoins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydantoin
- Alpha-amino acid or derivatives
- Nitroaromatic compound
- 2-nitrofuran
- Semicarbazone
- Dicarboximide
- Furan
- Heteroaromatic compound
- Semicarbazide
- C-nitro compound
- Carbonic acid derivative
- Organic nitro compound
- Allyl-type 1,3-dipolar organic compound
- Organic 1,3-dipolar compound
- Azacycle
- Oxacycle
- Propargyl-type 1,3-dipolar organic compound
- Carboxylic acid derivative
- Organic oxoazanium
- Carbonyl group
- Hydrocarbon derivative
- Organic nitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organonitrogen compound
- Organooxygen compound
- Organic zwitterion
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | Solid (1). |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 223-228°C | | Boiling Point | Not Available | | Solubility | 79.5 mg/L (at 24°C) | | LogP | -0.47 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dl-5910000000-28fa036f18e5a73e73b8 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0190000000-2f95bb616fd4cad3f729 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0390000000-c768fb1045d9566a1d4b | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9500000000-c45d8a5e217cd3045741 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-1090000000-f108972dd03c2012672f | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-b1f3423dbacafe623fb2 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-7491a417b52b7d6d99d2 | 2016-08-03 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-25 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Oral.
Readily absorbed in GI tract primarily in small intestine. Enhanced by food or delayed gastric emptying via enhanced dissolution rate of the drug. |
|---|
| Mechanism of Toxicity | Nitrofurantoin is activated by bacterial flavoproteins (nitrofuran reductase) to active reduced reactive intermediates that are thought to modulate and damage ribosomal proteins or other macromolecules, especially DNA, causing inhibition of DNA, RNA, protein, and cell wall synthesis. Nitrofurantoin inhibits bacterial acetyl-coenzyme A, interfering with the organism's carbohydrate metabolism. The drug also can disrupt bacterial cell wall formation. The overall effect is inhibition of bacterial growth or cell death. |
|---|
| Metabolism | Hepatic (75%) (2). Partially metabolized in liver to aminofurantoin.
Half Life: 0.3-1 hour |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | 3, not classifiable as to its carcinogenicity to humans. (5) |
|---|
| Uses/Sources | May be used as an alternative in the treatment of urinary tract infections. May be used by females pericoitally for prophylaxis against recurrent cystitis related to coitus. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Sialadenitis, pancreatitis, peripheral neuropathy, Asthenia, vertigo, and nystagmus, benign intracranial hypertension (pseudotumor cerebri), confusion, depression, optic neuritis, and psychotic reactions (1). |
|---|
| Symptoms | Acute toxicity may cause vomiting. Adverse effects include nausea and urine discolouration. Rare hepatotoxic and hypersensitivity reactions have occurred. Hemolytic anemia is a risk in patients with G6PD deficiency. Ascending polyneuropathy may occur with prolonged therapy or in patients with low creatinine clearance. |
|---|
| Treatment | There is no specific antidote, but a high fluid intake should be maintained to promote urinary excretion of the drug. Nitrofurantoin is dialyzable. (4) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00698 |
|---|
| HMDB ID | HMDB14836 |
|---|
| PubChem Compound ID | 5353830 |
|---|
| ChEMBL ID | CHEMBL572 |
|---|
| ChemSpider ID | 4510228 |
|---|
| KEGG ID | Not Available |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 1039679 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Nitrofurantoin |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Nitrofurantoin |
|---|
| References |
|---|
| Synthesis Reference | Tara Bielski, Kerry Benson, “Novel orally administrable formulation of nitrofurantoin and a method for preparing said formulation.” U.S. Patent US20050202079, issued September 15, 2005. |
|---|
| MSDS | Link |
|---|
| General References | - Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M: DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901-6. Epub 2007 Nov 29. [18048412 ]
- Wikipedia. Nitrofurantoin. Last Updated 8 August 2009. [Link]
- Drugs.com [Link]
- RxList: The Internet Drug Index (2009). [Link]
- International Agency for Research on Cancer (2014). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|