| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-21 20:27:32 UTC |
|---|
| Update Date | 2014-12-24 20:25:52 UTC |
|---|
| Accession Number | T3D2875 |
|---|
| Identification |
|---|
| Common Name | Riluzole |
|---|
| Class | Small Molecule |
|---|
| Description | Riluzole is only found in individuals that have used or taken this drug. It is a glutamate antagonist (receptors, glutamate) used as an anticonvulsant (anticonvulsants) and to prolong the survival of patients with amyotrophic lateral sclerosis. [PubChem]The mode of action of riluzole is unknown. Its pharmacological properties include the following, some of which may be related to its effect: 1) an inhibitory effect on glutamate release (activation of glutamate reuptake), 2) inactivation of voltage-dependent sodium channels, and 3) ability to interfere with intracellular events that follow transmitter binding at excitatory amino acid receptors. |
|---|
| Compound Type | - Amine
- Anesthetic
- Anticonvulsant
- Drug
- Ether
- Excitatory Amino Acid Antagonist
- Metabolite
- Neuroprotective Agent
- Organic Compound
- Organofluoride
- Synthetic Compound
|
|---|
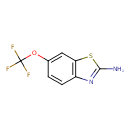
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | Fanizan | | Laidec | | Lizolorol | | Lizorolol | | Rilustad | | Rilutek | | Riluzol | | Riluzolum | | Sclefic | | Xie Yi Li | | Zolerilis |
|
|---|
| Chemical Formula | C8H5F3N2OS |
|---|
| Average Molecular Mass | 234.198 g/mol |
|---|
| Monoisotopic Mass | 234.007 g/mol |
|---|
| CAS Registry Number | 1744-22-5 |
|---|
| IUPAC Name | 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine |
|---|
| Traditional Name | riluzole |
|---|
| SMILES | FC(F)(F)OC1=CC2=C(NC(=N)S2)C=C1 |
|---|
| InChI Identifier | InChI=1S/C8H5F3N2OS/c9-8(10,11)14-4-1-2-5-6(3-4)15-7(12)13-5/h1-3H,(H2,12,13) |
|---|
| InChI Key | InChIKey=FTALBRSUTCGOEG-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as benzothiazoles. These are organic compounds containing a benzene fused to a thiazole ring (a five-membered ring with four carbon atoms, one nitrogen atom and one sulfur atom). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Benzothiazoles |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Benzothiazoles |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,3-benzothiazole
- Benzenoid
- 1,3-thiazol-2-amine
- Azole
- Thiazole
- Heteroaromatic compound
- Trihalomethane
- Azacycle
- Amine
- Organopnictogen compound
- Organic oxygen compound
- Alkyl halide
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organofluoride
- Organohalogen compound
- Organic nitrogen compound
- Alkyl fluoride
- Hydrocarbon derivative
- Halomethane
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 119°C | | Boiling Point | Not Available | | Solubility | 3.95e-02 g/L | | LogP | 2.3 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00li-2940000000-00c484cbc92e6a94bae0 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-2590000000-5a9d0f7cdad7e4f2ec6b | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-0390000000-4bd44ae3d25f047943e8 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-3940000000-1da38288b84e863a64e5 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-000i-0590000000-79d300cfd2b43692d963 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0190000000-63069bec4e00478eb380 | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0190000000-f0b481a2910122ad9bdc | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0090000000-d6a855ead1f5f5fe1a37 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-0900000000-b797bc65c27579651949 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-4caa5c429c103b56c151 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0090000000-78bd33541d87ef645303 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03di-0900000000-6f96de4ddfa58b872d2e | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0090000000-77cd58803ff632c89002 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0090000000-77cd58803ff632c89002 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4r-0490000000-ab117c07a1f16bb2d5f8 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-9f868134c3a763fd526e | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0090000000-9f868134c3a763fd526e | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9320000000-2312256d22d9ca57d014 | 2021-10-11 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Riluzole is well-absorbed (approximately 90%), with average absolute oral bioavailability of about 60% (CV=30%). A high fat meal decreases absorption, reducing AUC by about 20% and peak blood levels by about 45%. |
|---|
| Mechanism of Toxicity | The mode of action of riluzole is unknown. Its pharmacological properties include the following, some of which may be related to its effect: 1) an inhibitory effect on glutamate release (activation of glutamate reuptake), 2) inactivation of voltage-dependent sodium channels, and 3) ability to interfere with intracellular events that follow transmitter binding at excitatory amino acid receptors. |
|---|
| Metabolism | Riluzole is extensively metabolized to six major and a number of minor metabolites, which have not all been identified to date. Metabolism is mostly hepatic, consisting of cytochrome P450–dependent hydroxylation and glucuronidation. CYP1A2 is the primary isozyme involved in N-hydroxylation; CYP2D6, CYP2C19, CYP3A4, and CYP2E1 are considered unlikely to contribute significantly to riluzole metabolism in humans.
Half Life: The mean elimination half-life of riluzole is 12 hours (CV=35%) after repeated doses. |
|---|
| Toxicity Values | LD50: 85 mg/kg (p.o., mice) (8)
LD50: 34.5 mg/kg (i.v, mice) (8)
LD50: 45 mg/kg (p.o., rat) (8)
LD50: 21 mg/kg (i.v, mice) (8) |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | For the treatment of amyotrophic lateral sclerosis (ALS, Lou Gehrig's Disease) |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | May cause a potentially dangerous rash that may develop into Stevens Johnson syndrome, an extremely rare but potentially fatal skin disease. |
|---|
| Symptoms | Symptoms include such as nausea and fatigue [Wikipedia]. |
|---|
| Treatment | Treatment should be supportive and directed toward alleviating symptoms. Severe methemoglobinemia may be rapidly reversible after treatment with methylene blue. (9) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00740 |
|---|
| HMDB ID | HMDB14878 |
|---|
| PubChem Compound ID | 5070 |
|---|
| ChEMBL ID | CHEMBL744 |
|---|
| ChemSpider ID | 4892 |
|---|
| KEGG ID | C07937 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 138898 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Riluzole |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Riluzole |
|---|
| References |
|---|
| Synthesis Reference | Pratap Padi, Madhusudhan Ganta, Satyanarayana Bollikonda, Sridhar Chaganti, Ramulu Akula, Loka Maheshwari Dommati, “PROCESS FOR PREPARING RILUZOLE.” U.S. Patent US20080108827, issued May 08, 2008. |
|---|
| MSDS | T3D2875.pdf |
|---|
| General References | - Song JH, Huang CS, Nagata K, Yeh JZ, Narahashi T: Differential action of riluzole on tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels. J Pharmacol Exp Ther. 1997 Aug;282(2):707-14. [9262334 ]
- Coric V, Taskiran S, Pittenger C, Wasylink S, Mathalon DH, Valentine G, Saksa J, Wu YT, Gueorguieva R, Sanacora G, Malison RT, Krystal JH: Riluzole augmentation in treatment-resistant obsessive-compulsive disorder: an open-label trial. Biol Psychiatry. 2005 Sep 1;58(5):424-8. [15993857 ]
- van Kan HJ, Groeneveld GJ, Kalmijn S, Spieksma M, van den Berg LH, Guchelaar HJ: Association between CYP1A2 activity and riluzole clearance in patients with amyotrophic lateral sclerosis. Br J Clin Pharmacol. 2005 Mar;59(3):310-3. [15752377 ]
- Zarate CA Jr, Payne JL, Quiroz J, Sporn J, Denicoff KK, Luckenbaugh D, Charney DS, Manji HK: An open-label trial of riluzole in patients with treatment-resistant major depression. Am J Psychiatry. 2004 Jan;161(1):171-4. [14702270 ]
- Mathew SJ, Manji HK, Charney DS: Novel drugs and therapeutic targets for severe mood disorders. Neuropsychopharmacology. 2008 Aug;33(9):2080-92. doi: 10.1038/sj.npp.1301652. Epub 2008 Jan 2. [18172433 ]
- Lamanauskas N, Nistri A: Riluzole blocks persistent Na+ and Ca2+ currents and modulates release of glutamate via presynaptic NMDA receptors on neonatal rat hypoglossal motoneurons in vitro. Eur J Neurosci. 2008 May;27(10):2501-14. doi: 10.1111/j.1460-9568.2008.06211.x. Epub 2008 Apr 26. [18445055 ]
- Drugs.com [Link]
- Sanofi Aventis Fact Sheet for Rilutek(Riluzole) [Link]
- RxList: The Internet Drug Index (2009). [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | | Gene | Gene Symbol | Gene ID | Interaction | Chromosome | Details |
|---|
|
|---|
| Down-Regulated Genes | Not Available |
|---|