| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-30 17:59:02 UTC |
|---|
| Update Date | 2014-12-24 20:26:07 UTC |
|---|
| Accession Number | T3D3522 |
|---|
| Identification |
|---|
| Common Name | 1,1-Dimethylbiguanide |
|---|
| Class | Small Molecule |
|---|
| Description | Metformin is the most popular anti-diabetic drug in the United States and one of the most prescribed drugs in the country overall, with nearly 35 million prescriptions filled in 2006 for generic metformin alone. Metformin is a biguanide antihyperglycemic agent used for treating non-insulin-dependent diabetes mellitus (NIDDM). It improves glycemic control by decreasing hepatic glucose production, decreasing glucose absorption and increasing insulin-mediated glucose uptake. Metformin is the only oral antihyperglycemic agent that is not associated with weight gain. Metformin may induce weight loss and is the drug of choice for obese NIDDM patients. When used alone, metformin does not cause hypoglycemia; however, it may potentiate the hypoglycemic effects of sulfonylureas and insulin. Its main side effects are dyspepsia, nausea and diarrhea. Dose titration and/or use of smaller divided doses may decrease side effects. Metformin should be avoided in those with severely compromised renal function (creatinine clearance < 30 ml/min), acute/decompensated heart failure, severe liver disease and for 48 hours after the use of iodinated contrast dyes due to the risk of lactic acidosis. Lower doses should be used in the elderly and those with decreased renal function. Metformin decreases fasting plasma glucose, postprandial blood glucose and glycosolated hemoglobin (HbA1c) levels, which are reflective of the last 8-10 weeks of glucose control. Metformin may also have a positive effect on lipid levels. In 2012, a combination tablet of linagliptin plus metformin hydrochloride was marketed under the name Jentadueto for use in patients when treatment with both linagliptin and metformin is appropriate. |
|---|
| Compound Type | - Amide
- Amine
- Antidiabetic Agent
- Drug
- Food Toxin
- Hypoglycemic Agent
- Metabolite
- Organic Compound
- Synthetic Compound
|
|---|
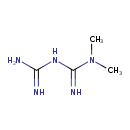
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 1,1-Dimethyl biguanide | | Apo-Metformin | | Diabefagos | | Diabex | | Diaformin | | Dianben | | Dimethylbiguanid | | Dimethylbiguanide | | DMBG | | Fortamet | | Gen-Metformin | | Glucophage | | Glucophage XR | | Glumetza | | Haurymellin | | Meguan | | Metformin | | Metformina | | Metformine | | Metformine pamoate | | Metforminum | | Metiguanide | | Mylan-Metformin | | N,N-Dimethylbiguanide | | N,N-Dimethylimidodicarbonimidic diamide | | N1,N1-Dimethylbiguanide | | Novo-Metformin | | Nu-Metformin | | Obimet | | PMS-Metformin | | Ran-Metformin | | Ratio-Metformin | | Riomet | | Sandoz Metformin | | Teva-Metformin |
|
|---|
| Chemical Formula | C4H11N5 |
|---|
| Average Molecular Mass | 129.164 g/mol |
|---|
| Monoisotopic Mass | 129.101 g/mol |
|---|
| CAS Registry Number | 657-24-9 |
|---|
| IUPAC Name | 1-carbamimidamido-N,N-dimethylmethanimidamide |
|---|
| Traditional Name | metformin |
|---|
| SMILES | CN(C)C(=N)NC(N)=N |
|---|
| InChI Identifier | InChI=1S/C4H11N5/c1-9(2)4(7)8-3(5)6/h1-2H3,(H5,5,6,7,8) |
|---|
| InChI Key | InChIKey=XZWYZXLIPXDOLR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as biguanides. These are organic compounds containing two N-linked guanidines. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Guanidines |
|---|
| Direct Parent | Biguanides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Biguanide
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carboximidamide
- Organopnictogen compound
- Hydrocarbon derivative
- Imine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | - Erythrocyte
- Liver
- Muscle
- Skeletal Muscle
|
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 223-226°C | | Boiling Point | Not Available | | Solubility | Freely soluble as HCl salt | | LogP | -0.5 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9100000000-4cf43a0a5a8d4db19a46 | 2017-08-28 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-0900000000-9046e2aa0408a0396007 | 2012-07-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-01qi-9700000000-a6b98d87cc840a082179 | 2012-07-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dr-9000000000-8e80f301bad045540477 | 2012-07-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-001i-0900000000-bd8aed328c944acd1270 | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-03l9-9300000000-3d585674ffe84238e5bf | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-00di-9000000000-ee68820579ebe4d31082 | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-00di-9000000000-4312e7e5e1b0dd9ef936 | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-00di-9000000000-053d63fe09a95fc1d544 | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0229-9100000000-7fe999a9d1aaae3bbe53 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-001i-0900000000-c235cd5d0dda3f3c28d9 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-001i-0900000000-0fa445716bfc24131a75 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-3900000000-dee37da326e6f0b2c56a | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0900000000-45bd1f8c6d2dc4f38944 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-1900000000-38f3dedb5c19900cdefb | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-7900000000-bf5d1092aa372c303d61 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-03l9-9300000000-06a99f0dff4b41a23cfa | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-022i-9100000000-811c9e7cf8b30b27c0f2 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00di-9000000000-d34b9b3ab9eb78317eba | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0900000000-4d53ac0f7dfaf860e784 | 2017-09-14 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-6900000000-03a99ea4f96636a6d5e2 | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0079-9000000000-24ee057ec9505ccf8d3d | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0229-9000000000-bc7faff99aaa83f5e600 | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9300000000-e145cf58ec0f6be503ab | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9100000000-0cc6d6ceac8192964ee4 | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-f47eab8fa0fd4177cfdd | 2017-07-26 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | Not Available | 2012-12-05 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | 2012-12-05 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Absorbed over 6 hours, bioavailability is 50 to 60% under fasting conditions. Administration with food decreases and delays absorption. Some evidence indicates that the level of absorption is not dose-related, suggesting that absorption occurs through a saturable process. Limited data from animal and human cell cultures indicate that absorption occurs through a passive, non-saturable process, possibly involving a paracellular route. Peak action occurs 3 hours after oral administration. |
|---|
| Mechanism of Toxicity | Metformin's mechanisms of action differ from other classes of oral antihyperglycemic agents. Metformin decreases blood glucose levels by decreasing hepatic glucose production, decreasing intestinal absorption of glucose, and improving insulin sensitivity by increasing peripheral glucose uptake and utilization. These effects are mediated by the initial activation by metformin of AMP-activated protein kinase (AMPK), a liver enzyme that plays an important role in insulin signaling, whole body energy balance, and the metabolism of glucose and fats. Activation of AMPK is required for metformin's inhibitory effect on the production of glucose by liver cells. Increased peripheral utilization of glucose may be due to improved insulin binding to insulin receptors. Metformin administration also increases AMPK activity in skeletal muscle. AMPK is known to cause GLUT4 deployment to the plasma membrane, resulting in insulin-independent glucose uptake. The rare side effect, lactic acidosis, is thought to be caused by decreased liver uptake of serum lactate, one of the substrates of gluconeogenesis. In those with healthy renal function, the slight excess is simply cleared. However, those with severe renal impairment may accumulate clinically significant serum lactic acid levels. Other conditions that may precipitate lactic acidosis include severe hepatic disease and acute/decompensated heart failure.
|
|---|
| Metabolism | Metformin is not metabolized.
Route of Elimination: Intravenous single-dose studies in normal subjects demonstrate that metformin is excreted unchanged in the urine and does not undergo hepatic metabolism (no metabolites have been identified in humans) nor biliary excretion. Approximately 90% of the drug is eliminated in 24 hours in those with healthy renal function. Renal clearance of metformin is approximately 3.5 times that of creatinine clearance, indicating the tubular secretion is the primary mode of metformin elimination.
Half Life: 6.2 hours. Duration of action is 8-12 hours. |
|---|
| Toxicity Values | Acute oral toxicity (LD50): 350 mg/kg [Rabbit]. |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | For use as an adjunct to diet and exercise in adult patients (18 years and older) with NIDDM. May also be used for the management of metabolic and reproductive abnormalities associated with polycystic ovary syndrome (PCOS). Jentadueto is for the treatment of patients when both linagliptin and metformin is appropriate. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Acute oral toxicity (LD50): 350 mg/kg [Rabbit]. It would be expected that adverse reactions of a more intense character including epigastric discomfort, nausea, and vomiting followed by diarrhea, drowsiness, weakness, dizziness, malaise and headache might be seen. |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00331 |
|---|
| HMDB ID | HMDB01921 |
|---|
| PubChem Compound ID | 4091 |
|---|
| ChEMBL ID | CHEMBL1431 |
|---|
| ChemSpider ID | 3949 |
|---|
| KEGG ID | C07151 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | 125853 , 184700 |
|---|
| ChEBI ID | 6801 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Metformin |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Metformin |
|---|
| References |
|---|
| Synthesis Reference | Jorn Moeckel, Rolf-Dieter Gabel, Heinrich Woog, “Pharmaceutical preparation containing metformin and a process for producing it.” U.S. Patent US5955106, issued October, 1991. |
|---|
| MSDS | Link |
|---|
| General References | - Witters LA: The blooming of the French lilac. J Clin Invest. 2001 Oct;108(8):1105-7. [11602616 ]
- UNGAR G, FREEDMAN L, SHAPIRO SL: Pharmacological studies of a new oral hypoglycemic drug. Proc Soc Exp Biol Med. 1957 May;95(1):190-2. [13432032 ]
- Lord JM, Flight IH, Norman RJ: Metformin in polycystic ovary syndrome: systematic review and meta-analysis. BMJ. 2003 Oct 25;327(7421):951-3. [14576245 ]
- Marchesini G, Brizi M, Bianchi G, Tomassetti S, Zoli M, Melchionda N: Metformin in non-alcoholic steatohepatitis. Lancet. 2001 Sep 15;358(9285):893-4. [11567710 ]
- Nair S, Diehl AM, Wiseman M, Farr GH Jr, Perrillo RP: Metformin in the treatment of non-alcoholic steatohepatitis: a pilot open label trial. Aliment Pharmacol Ther. 2004 Jul 1;20(1):23-8. [15225167 ]
- Seale FG 4th, Robinson RD, Neal GS: Association of metformin and pregnancy in the polycystic ovary syndrome. A report of three cases. J Reprod Med. 2000 Jun;45(6):507-10. [10900588 ]
- Briggs GG, Ambrose PJ, Nageotte MP, Padilla G, Wan S: Excretion of metformin into breast milk and the effect on nursing infants. Obstet Gynecol. 2005 Jun;105(6):1437-41. [15932841 ]
- De Jager J, Kooy A, Lehert P, Bets D, Wulffele MG, Teerlink T, Scheffer PG, Schalkwijk CG, Donker AJ, Stehouwer CD: Effects of short-term treatment with metformin on markers of endothelial function and inflammatory activity in type 2 diabetes mellitus: a randomized, placebo-controlled trial. J Intern Med. 2005 Jan;257(1):100-9. [15606381 ]
- Mughal MA, Jan M, Maheri WM, Memon MY, Ali M: The effect of metformin on glycemic control, serum lipids and lipoproteins in diet alone and sulfonylurea-treated type 2 diabetic patients with sub-optimal metabolic control. J Pak Med Assoc. 2000 Nov;50(11):381-6. [11126815 ]
- Einhorn D, Rendell M, Rosenzweig J, Egan JW, Mathisen AL, Schneider RL: Pioglitazone hydrochloride in combination with metformin in the treatment of type 2 diabetes mellitus: a randomized, placebo-controlled study. The Pioglitazone 027 Study Group. Clin Ther. 2000 Dec;22(12):1395-409. [11192132 ]
- Robert F, Fendri S, Hary L, Lacroix C, Andrejak M, Lalau JD: Kinetics of plasma and erythrocyte metformin after acute administration in healthy subjects. Diabetes Metab. 2003 Jun;29(3):279-83. [12909816 ]
- Radziuk J, Pye S: Hepatic glucose uptake, gluconeogenesis and the regulation of glycogen synthesis. Diabetes Metab Res Rev. 2001 Jul-Aug;17(4):250-72. [11544610 ]
- Morin-Papunen LC, Vauhkonen I, Koivunen RM, Ruokonen A, Martikainen HK, Tapanainen JS: Endocrine and metabolic effects of metformin versus ethinyl estradiol-cyproterone acetate in obese women with polycystic ovary syndrome: a randomized study. J Clin Endocrinol Metab. 2000 Sep;85(9):3161-8. [10999803 ]
- Musi N, Hirshman MF, Nygren J, Svanfeldt M, Bavenholm P, Rooyackers O, Zhou G, Williamson JM, Ljunqvist O, Efendic S, Moller DE, Thorell A, Goodyear LJ: Metformin increases AMP-activated protein kinase activity in skeletal muscle of subjects with type 2 diabetes. Diabetes. 2002 Jul;51(7):2074-81. [12086935 ]
- Marathe PH, Wen Y, Norton J, Greene DS, Barbhaiya RH, Wilding IR: Effect of altered gastric emptying and gastrointestinal motility on metformin absorption. Br J Clin Pharmacol. 2000 Oct;50(4):325-32. [11012555 ]
- Gore DC, Wolf SE, Sanford A, Herndon DN, Wolfe RR: Influence of metformin on glucose intolerance and muscle catabolism following severe burn injury. Ann Surg. 2005 Feb;241(2):334-42. [15650645 ]
- Gillies PS, Dunn CJ: Pioglitazone. Drugs. 2000 Aug;60(2):333-43; discussion 344-5. [10983737 ]
- Hale TW, Kristensen JH, Hackett LP, Kohan R, Ilett KF: Transfer of metformin into human milk. Diabetologia. 2002 Nov;45(11):1509-14. Epub 2002 Sep 25. [12436333 ]
- Lalau JD, Lacroix C: Measurement of metformin concentration in erythrocytes: clinical implications. Diabetes Obes Metab. 2003 Mar;5(2):93-8. [12630933 ]
- Kouki T, Takasu N, Nakachi A, Tamanaha T, Komiya I, Tawata M: Low-dose metformin improves hyperglycaemia related to myotonic dystrophy. Diabet Med. 2005 Mar;22(3):346-7. [15717887 ]
- Magalhaes FO, Gouveia LM, Torquato MT, Paccola GM, Piccinato CE, Foss MC: Metformin increases blood flow and forearm glucose uptake in a group of non-obese type 2 diabetes patients. Horm Metab Res. 2006 Aug;38(8):513-7. [16941277 ]
- Gore DC, Herndon DN, Wolfe RR: Comparison of peripheral metabolic effects of insulin and metformin following severe burn injury. J Trauma. 2005 Aug;59(2):316-22; discussion 322-3. [16294070 ]
- Bridger T, MacDonald S, Baltzer F, Rodd C: Randomized placebo-controlled trial of metformin for adolescents with polycystic ovary syndrome. Arch Pediatr Adolesc Med. 2006 Mar;160(3):241-6. [16520442 ]
- Amador-Licona N, Guizar-Mendoza J, Vargas E, Sanchez-Camargo G, Zamora-Mata L: The short-term effect of a switch from glibenclamide to metformin on blood pressure and microalbuminuria in patients with type 2 diabetes mellitus. Arch Med Res. 2000 Nov-Dec;31(6):571-5. [11257323 ]
- Carter D, Howlett HC, Wiernsperger NF, Bailey C: Effects of metformin on bile salt transport by monolayers of human intestinal Caco-2 cells. Diabetes Obes Metab. 2002 Nov;4(6):424-7. [12406042 ]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | | Gene | Gene Symbol | Gene ID | Interaction | Chromosome | Details |
|---|
|
|---|
| Down-Regulated Genes | | Gene | Gene Symbol | Gene ID | Interaction | Chromosome | Details |
|---|
|
|---|