| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2013-04-25 07:56:54 UTC |
|---|
| Update Date | 2014-12-24 20:26:34 UTC |
|---|
| Accession Number | T3D3913 |
|---|
| Identification |
|---|
| Common Name | Pymetrozine |
|---|
| Class | Small Molecule |
|---|
| Description | Pymetrozine is a neuroactive insecticide that selectively affects chordotonal mechanoreceptors. Physiologically, it appears to act by preventing these insects from inserting their stylus into the plant tissue. It controls many strains of aphids on potatoes and in a range of vegetable brassica crops. Pymetrozine has been determined to be of low acute toxicity to humans, birds, aquatic organisms, mammals, and bees. |
|---|
| Compound Type | - Insecticide
- Organic Compound
- Pesticide
- Synthetic Compound
|
|---|
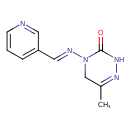
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (e)-4,5-dihydro-6-Methyl-4-((3-pyridinylmethylene)amino)-1,2,4-triazin-3(2H)-one |
|
|---|
| Chemical Formula | C10H11N5O |
|---|
| Average Molecular Mass | 217.227 g/mol |
|---|
| Monoisotopic Mass | 217.096 g/mol |
|---|
| CAS Registry Number | 123312-89-0 |
|---|
| IUPAC Name | 6-methyl-4-[(E)-(pyridin-3-ylmethylidene)amino]-2,3,4,5-tetrahydro-1,2,4-triazin-3-one |
|---|
| Traditional Name | endeavor |
|---|
| SMILES | CC1=NNC(=O)N(C1)\N=C\C1=CC=CN=C1 |
|---|
| InChI Identifier | InChI=1S/C10H11N5O/c1-8-7-15(10(16)14-13-8)12-6-9-3-2-4-11-5-9/h2-6H,7H2,1H3,(H,14,16)/b12-6+ |
|---|
| InChI Key | InChIKey=QHMTXANCGGJZRX-WUXMJOGZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1,2,4-triazines. 1,2,4-triazines are compounds containing a triazine ring, which is a heterocyclic ring, similar to the six-member benzene ring but with three carbons replaced by nitrogen atoms, at ring positions 1, 2, and 4. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Triazines |
|---|
| Sub Class | 1,2,4-triazines |
|---|
| Direct Parent | 1,2,4-triazines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1,2,4-triazine
- Pyridine
- Heteroaromatic compound
- Azacycle
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0290000000-d0ee4d64fc31b113c278 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dr-1910000000-ead59c3a233a73eb19e1 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053u-9300000000-bec7727375c76f341c5f | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-9730000000-8cf9aa6f3aa48ccd9a1e | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0007-9400000000-249d0b9ca3daad2e9b64 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-0d62d3658487b30bb886 | 2016-08-03 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-03dj-9300000000-0227c62ae652df68c9c2 | 2014-10-20 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, DMSO-d6, experimental) | Not Available | 2014-10-20 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100.40 MHz, DMSO-d6, experimental) | Not Available | 2014-10-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | This is a man-made compound that is used as a pesticide. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 9576037 |
|---|
| ChEMBL ID | CHEMBL1906026 |
|---|
| ChemSpider ID | 7850487 |
|---|
| KEGG ID | C18590 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 39311 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D3913.pdf |
|---|
| General References | Not Available |
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|