| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-09-11 05:15:34 UTC |
|---|
| Update Date | 2014-12-24 20:26:56 UTC |
|---|
| Accession Number | T3D4764 |
|---|
| Identification |
|---|
| Common Name | Erythromycin |
|---|
| Class | Small Molecule |
|---|
| Description | Erythromycin is a macrolide antibiotic produced by Streptomyces erythreus. It inhibits bacterial protein synthesis by binding to bacterial 50S ribosomal subunits; binding inhibits peptidyl transferase activity and interferes with translocation of amino acids during translation and assembly of proteins. Erythromycin may be bacteriostatic or bactericidal depending on the organism and drug concentration. |
|---|
| Compound Type | - Amine
- Anti-Bacterial Agent
- Drug
- Ester
- Ether
- Gastrointestinal Agent
- Macrolide
- Metabolite
- Organic Compound
- Protein Synthesis Inhibitor
- Synthetic Compound
|
|---|
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 3''-O-demethylerythromycin | | Abomacetin | | Akne-Mycin | | E.E.S | | E.E.S. | | EM | | Eritromicina | | Ery | | Ery-Ped | | ERY-TAB | | Eryc | | Erygel | | Erythra-Derm | | Erythrocin Stearate | | Erythromycin A | | Erythromycin C | | Erythromycin ethylsuccinate | | Erythromycin glucoheptonate | | Erythromycin lactobionate | | Erythromycin oxime | | Erythromycin Stearate | | Erythromycine | | Erythromycinum | | Ilosone | | ILOTYCIN | | Staticin | | T-stat |
|
|---|
| Chemical Formula | C37H67NO13 |
|---|
| Average Molecular Mass | 733.927 g/mol |
|---|
| Monoisotopic Mass | 733.461 g/mol |
|---|
| CAS Registry Number | 114-07-8 |
|---|
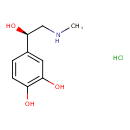
| IUPAC Name | 4-[(1R)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol hydrochloride |
|---|
| Traditional Name | epinephrine hydrochloride |
|---|
| SMILES | [H][C@@]1(C)C[C@]([H])(N(C)C)[C@@]([H])(O)[C@]([H])(O[C@]2([H])[C@@]([H])(C)[C@]([H])(O[C@@]3([H])C[C@@](C)(OC)[C@@]([H])(O)[C@]([H])(C)O3)[C@@]([H])(C)C(=O)O[C@]([H])(CC)[C@@](C)(O)[C@]([H])(O)[C@@]([H])(C)C(=O)[C@]([H])(C)C[C@@]2(C)O)O1 |
|---|
| InChI Identifier | InChI=1S/C37H67NO13/c1-14-25-37(10,45)30(41)20(4)27(39)18(2)16-35(8,44)32(51-34-28(40)24(38(11)12)15-19(3)47-34)21(5)29(22(6)33(43)49-25)50-26-17-36(9,46-13)31(42)23(7)48-26/h18-26,28-32,34,40-42,44-45H,14-17H2,1-13H3/t18-,19-,20+,21+,22-,23+,24+,25-,26+,28-,29+,30-,31+,32-,34+,35-,36-,37-/m1/s1 |
|---|
| InChI Key | InChIKey=ULGZDMOVFRHVEP-RWJQBGPGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as catechols. Catechols are compounds containing a 1,2-benzenediol moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Phenols |
|---|
| Sub Class | Benzenediols |
|---|
| Direct Parent | Catechols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Aralkylamine
- Monocyclic benzene moiety
- 1,2-aminoalcohol
- Secondary alcohol
- Secondary aliphatic amine
- Secondary amine
- Alcohol
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Aromatic alcohol
- Hydrochloride
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 191°C | | Boiling Point | Not Available | | Solubility | 2000mg/L at 28°C | | LogP | 3.06 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - DI-ESI-Ion Trap , Positive | splash10-00di-2901000023-e1b4c2a4d54a44d851b1 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - DI-ESI-Hybrid FT , Positive | splash10-00di-2901000023-e1b4c2a4d54a44d851b1 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a7i-0900070700-45f1a3bf3aac26069507 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a4i-0900020000-c282d6a703a3a812c91a | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a4i-0900000000-b11e26802dca28ea88f0 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0a4i-0900000000-5ea1e94063a49c0d6ddb | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-004i-0000090000-f0fcdc5ed4d00170f2a3 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-0200000900-439b7d31b57ea93dff40 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0a59-4900000000-1cb488a53f50cba638cf | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9700000000-239fd7095409362c75bc | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9000000000-37757a9a6c61ec2bac1b | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9000000000-2d04ae69b21d82ee0763 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9000000000-2d04ae69b21d82ee0763 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-053r-0500000900-20a1c46dfdef404c9d3e | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0a59-5900000000-c7e9166d7e79e23023a5 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-053r-9800000000-07e4c66bcd0be93f7c09 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9200000000-553fb27002eceacb9270 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9000000000-9725c29b26f7cbbef576 | 2018-05-25 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-001i-9000000000-2d04ae69b21d82ee0763 | 2018-05-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0090000000-4c8f943d869f13c9bb1c | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0090000000-4c8f943d869f13c9bb1c | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-0090000000-4c8f943d869f13c9bb1c | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0090000000-1a30917d4f392e75cfd3 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0090000000-1a30917d4f392e75cfd3 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-014i-0090000000-1a30917d4f392e75cfd3 | 2016-08-03 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Orally administered erythromycin base and its salts are readily absorbed in the microbiologically active form. Topical application of the ophthalmic ointment to the eye may result in absorption into the cornea and aqueous humor. |
|---|
| Mechanism of Toxicity | Erythromycin acts by penetrating the bacterial cell membrane and reversibly binding to the 50 S subunit of bacterial ribosomes or near the “P” or donor site so that binding of tRNA (transfer RNA) to the donor site is blocked. Translocation of peptides from the “A” or acceptor site to the “P” or donor site is prevented, and subsequent protein synthesis is inhibited. Erythromycin is effective only against actively dividing organisms. The exact mechanism by which erythmromycin reduces lesions of acne vulgaris is not fully known: however, the effect appears to be due in part to the antibacterial activity of the drug. |
|---|
| Metabolism | Hepatic. Extensively metabolized - after oral administration, less than 5% of the administered dose can be recovered in the active form in the urine. Erythromycin is partially metabolized by CYP3A4 resulting in numerous drug interactions.
Half Life: 0.8 - 3 hours |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | For use in the treatment of infections caused by susceptible strains of microorganisms in the following diseases: respiratory tract infections (upper and lower) of mild to moderate degree, pertussis (whooping cough), as adjunct to antitoxin in infections due to Corynebacterium diphtheriae, in the treatment of infections due to Corynebacterium minutissimum, intestinal amebiasis caused by Entamoeba histolytica, acute pelvic inflammatory disease caused by Neisseria gonorrhoeae, skin and soft tissue infections of mild to moderate severity caused by Streptococcus pyogenes and Staphylococcus aureus, primary syphilis caused by Treponema pallidum, infections caused by Chlamydia trachomatis, nongonococcal urethritis caused by Ureaplasma urealyticum, and Legionnaires' disease caused by Legionella pneumophila. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Symptoms of overdose include diarrhea, nausea, stomach cramps, and vomiting. |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00199 |
|---|
| HMDB ID | HMDB14344 |
|---|
| PubChem Compound ID | 12560 |
|---|
| ChEMBL ID | CHEMBL532 |
|---|
| ChemSpider ID | 12041 |
|---|
| KEGG ID | C01912 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 48923 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | ERY |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Erythromycin |
|---|
| References |
|---|
| Synthesis Reference | Takehiro Amano, Masami Goi, Kazuto Sekiuchi, Tomomichi Yoshida, Masahiro Hasegawa, “Process for preparing erythromycin A oxime or a salt thereof.” U.S. Patent US5274085, issued October, 1966. |
|---|
| MSDS | Link |
|---|
| General References | - Kanazawa S, Ohkubo T, Sugawara K: The effects of grapefruit juice on the pharmacokinetics of erythromycin. Eur J Clin Pharmacol. 2001 Jan-Feb;56(11):799-803. [11294369 ]
- Ogwal S, Xide TU: Bioavailability and stability of erythromycin delayed release tablets. Afr Health Sci. 2001 Dec;1(2):90-6. [12789122 ]
- Okudaira T, Kotegawa T, Imai H, Tsutsumi K, Nakano S, Ohashi K: Effect of the treatment period with erythromycin on cytochrome P450 3A activity in humans. J Clin Pharmacol. 2007 Jul;47(7):871-6. [17585116 ]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|