| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2009-07-21 20:26:36 UTC |
|---|
| Update Date | 2014-12-24 20:25:51 UTC |
|---|
| Accession Number | T3D2752 |
|---|
| Identification |
|---|
| Common Name | Hydromorphone |
|---|
| Class | Small Molecule |
|---|
| Description | Hydromorphone is only found in individuals that have used or taken this drug. It is an opioid analgesic derived from morphine and used mainly as an analgesic. It has a shorter duration of action and is more potent than morphine. Hydromorphone is a narcotic analgesic; its principal therapeutic effect is relief of pain. Hydromorphone interacts predominantly with the opioid mu-receptors. These mu-binding sites are discretely distributed in the human brain, with high densities in the posterior amygdala, hypothalamus, thalamus, nucleus caudatus, putamen, and certain cortical areas. They are also found on the terminal axons of primary afferents within laminae I and II (substantia gelatinosa) of the spinal cord and in the spinal nucleus of the trigeminal nerve. In clinical settings, Hydromorphone exerts its principal pharmacological effect on the central nervous system and gastrointestinal tract. Hydromorphone also binds with kappa-receptors which are thought to mediate spinal analgesia, miosis and sedation. |
|---|
| Compound Type | - Amine
- Analgesic, Opioid
- Drug
- Ether
- Metabolite
- Narcotic
- Organic Compound
- Synthetic Compound
|
|---|
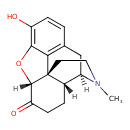
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (-)-(5R)-4,5-Epoxy-3-hydroxy-9alpha-methylmorphinan-6-one | | 4,5-Epoxy-3-hydroxy-17-methylmorphinan-6-one | | 4,5alpha-Epoxy-3-hydroxy-17-methyl-6-morphinanone | | 6-Deoxy-7,8-dihydro-6-oxomorphine | | 7,8-Dihydromorphinone | | Dihydromorfinon | | Dihydromorphinone | | Dilaudid | | Dimorphone | | EXALGO | | Hidromorfona | | Hydromorfona | | Hydromorphon | | Hydromorphonum | | Idromorfone | | Palladone |
|
|---|
| Chemical Formula | C17H19NO3 |
|---|
| Average Molecular Mass | 285.338 g/mol |
|---|
| Monoisotopic Mass | 285.136 g/mol |
|---|
| CAS Registry Number | 466-99-9 |
|---|
| IUPAC Name | (1S,5R,13R,17R)-10-hydroxy-4-methyl-12-oxa-4-azapentacyclo[9.6.1.0¹,¹³.0⁵,¹⁷.0⁷,¹⁸]octadeca-7(18),8,10-trien-14-one |
|---|
| Traditional Name | hydromorphone |
|---|
| SMILES | [H][C@@]12OC3=C(O)C=CC4=C3[C@@]11CCN(C)[C@]([H])(C4)[C@]1([H])CCC2=O |
|---|
| InChI Identifier | InChI=1S/C17H19NO3/c1-18-7-6-17-10-3-5-13(20)16(17)21-15-12(19)4-2-9(14(15)17)8-11(10)18/h2,4,10-11,16,19H,3,5-8H2,1H3/t10-,11+,16-,17-/m0/s1 |
|---|
| InChI Key | InChIKey=WVLOADHCBXTIJK-YNHQPCIGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as morphinans. These are polycyclic compounds with a four-ring skeleton with three condensed six-member rings forming a partially hydrogenated phenanthrene moiety, one of which is aromatic while the two others are alicyclic. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Alkaloids and derivatives |
|---|
| Class | Morphinans |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Morphinans |
|---|
| Alternative Parents | |
|---|
| Substituents | - Morphinan
- Phenanthrene
- Isoquinolone
- Tetralin
- Coumaran
- Alkyl aryl ether
- 1-hydroxy-2-unsubstituted benzenoid
- Aralkylamine
- Piperidine
- Benzenoid
- Ketone
- Tertiary amine
- Tertiary aliphatic amine
- Oxacycle
- Ether
- Azacycle
- Organoheterocyclic compound
- Amine
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Carbonyl group
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane
|
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 266.5°C | | Boiling Point | Not Available | | Solubility | 4.39e+00 g/L | | LogP | 0.9 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0adl-2090000000-1b72d2e162be578cb94d | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-006x-6039000000-d29846cb85959ad1cc2c | 2017-10-06 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-000i-0290000000-0c258477ac267eb9243c | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0090000000-052fc44835772dd50b65 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0090000000-711917845b2129c2d3e3 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-7090000000-8d20bdd275a62a96fd87 | 2016-06-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-a0979bf03853322bb9f7 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0090000000-2cd300544d508b429bee | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00ou-1290000000-c40fe8399a0aae44bc33 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0090000000-12d6d7cf9e71a6a9ec09 | 2021-09-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0090000000-94ce4b4a1e2ea08a7fc9 | 2021-09-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-2090000000-aa4c1402684fd8359729 | 2021-09-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0090000000-793f02ff50658d83401d | 2021-09-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0090000000-c9bb75a9e6ed1373a730 | 2021-09-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0090000000-4e4413edce04e0b80bd6 | 2021-09-25 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-002r-6960000000-9f3d9b17d56e032d76c5 | 2014-09-20 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Parental (intravenous, intramuscular); oral; enteral(rectal).
Better absorbed orally than morphine. |
|---|
| Mechanism of Toxicity | Hydromorphone is a narcotic analgesic; its principal therapeutic effect is relief of pain. Hydromorphone interacts predominantly with the opioid mu-receptors. These mu-binding sites are discretely distributed in the human brain, with high densities in the posterior amygdala, hypothalamus, thalamus, nucleus caudatus, putamen, and certain cortical areas. They are also found on the terminal axons of primary afferents within laminae I and II (substantia gelatinosa) of the spinal cord and in the spinal nucleus of the trigeminal nerve. In clinical settings, Hydromorphone exerts its principal pharmacological effect on the central nervous system and gastrointestinal tract. Hydromorphone also binds with kappa-receptors which are thought to mediate spinal analgesia, miosis and sedation. |
|---|
| Metabolism | Primarily hepatic. After absorption hydromorphone is metabolized by the liver to the glucuronide conjugate which is then excreted in the urine. Hydromorphone is metabolized to the major metabolites hydromorphone-3-glucuronide, hydromorphone-3-glucoside and dihydroisomorphine-6-glucuronide.
Route of Elimination: Only a small amount of the hydromorphone dose is excreted unchanged in the urine.
Most of the dose is excreted as hydromorphone-3-glucuronide along with minor amounts of 6-hydroxy reduction metabolites.
Half Life: 2.6 hours (oral); 18.6 hours for sustained release Palladone |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | For the relief of moderate to severe pain such as that due to surgery, cancer, trauma/injury, or burns. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Medical problems can include congested lungs, liver disease, tetanus, infection of the heart valves, skin abscesses, anemia and pneumonia. Death can occur from overdose. |
|---|
| Symptoms | Hydromorphone is a schedule II narcotic which can lead to physical dependence or addiction. High doses lead to respiratory depression, nausea, and vomiting. Overdoses lead to extreme somnolence progressing to stupor or coma, skeletal muscle flaccidity, cold and clammy skin, and sometimes bradycardia and hypotension. In severe overdosage, apnea, circulatory collapse, cardiac arrest and death may occur. |
|---|
| Treatment | In the treatment of overdosage, primary attention should be given to the reestablishment of adequate respiratory exchange through provision of a patent airway and institution of assisted or controlled ventilation. A potentially serious oral ingestion, if recent, should be managed with gut decontamination. In unconscious patients with a secure airway, instill activated charcoal (30-100 g in adults, 1-2 g/kg in infants) via a nasogastric tube. A saline cathartic or sorbitol may be added to the first dose of activated charcoal. Supportive measures (including oxygen, vasopressors) should be employed in the management of circulatory shock and pulmonary edema accompanying overdose as indicated. Cardiac arrest or arrhythmias may require cardiac massage or defibrillation. The opioid antagonist, naloxone, is a specific antidote against respiratory depression which may result from overdosage, or unusual sensitivity to Hydromorphone. Therefore, an appropriate dose of this antagonist should be administered, preferably by the intravenous route, simultaneously with efforts at respiratory resuscitation. Naloxone should not be administered in the absence of clinically significant respiratory or circulatory depression. (5) |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00327 |
|---|
| HMDB ID | HMDB14472 |
|---|
| PubChem Compound ID | 5284570 |
|---|
| ChEMBL ID | CHEMBL398707 |
|---|
| ChemSpider ID | 4447624 |
|---|
| KEGG ID | C07042 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 5790 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Hydromorphone |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Hydromorphone |
|---|
| References |
|---|
| Synthesis Reference | Anne M. Hailes, Christopher E. French, Neil C. Bruce, “Morphinone reductase for the preparation of hydromorphone and hydrocodone.” U.S. Patent US5571685, issued November, 1990. |
|---|
| MSDS | Link |
|---|
| General References | - Coda BA, Rudy AC, Archer SM, Wermeling DP: Pharmacokinetics and bioavailability of single-dose intranasal hydromorphone hydrochloride in healthy volunteers. Anesth Analg. 2003 Jul;97(1):117-23, table of contents. [12818953 ]
- Vallner JJ, Stewart JT, Kotzan JA, Kirsten EB, Honigberg IL: Pharmacokinetics and bioavailability of hydromorphone following intravenous and oral administration to human subjects. J Clin Pharmacol. 1981 Apr;21(4):152-6. [6165742 ]
- Drugs.com [Link]
- Drugs.com [Link]
- RxList: The Internet Drug Index (2009). [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|