Flumioxazin (T3D3847)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2013-04-25 07:56:51 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:33 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D3847 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Flumioxazin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Flumioxazin is a broad-spectrum contact herbicide. It works by interfering with the plants’ production of chlorophyll. Treated plants will respond quickly to treatment and rapidly decompose. Flumioxazin is an N-phenylphthalimide herbicide. The mode of action in this family of herbicides is believed to be inhibition of protoporphyrinogen oxidase, an enzyme important in the synthesis of chlorophyll. Mechanistic study findings suggest that porphyrins accumulate in susceptible plants causing photosensitization, which leads to membrane lipid peroxidation. The peroxidation of membrane lipids leads to irreversible damage of membrane function and structure in susceptible plants. Treatment of soil with flumioxazin will cause susceptible emerging plants to turn necrotic and die shortly after exposure to sunlight. Flumioxazin is used on soybeans, peanuts, orchard fruits and other crops. Flumioxazin is also used to controls invasive Eurasian watermilfoil (Myriophyllum spicatum) and curlyleaf pondweed (Potamogeton crispus). Flumioxazin has been used as an agricultural chemical since 2001, and was conditionally registered for aquatic use in 2010. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
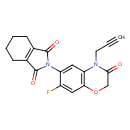
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C19H15FN2O4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 354.332 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 354.102 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 103361-09-7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 2-[7-fluoro-3-oxo-4-(prop-2-yn-1-yl)-3,4-dihydro-2H-1,4-benzoxazin-6-yl]-2,3,4,5,6,7-hexahydro-1H-isoindole-1,3-dione | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | flumioxazin | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | FC1=CC2=C(C=C1N1C(=O)C3=C(CCCC3)C1=O)N(CC#C)C(=O)CO2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C19H15FN2O4/c1-2-7-21-15-9-14(13(20)8-16(15)26-10-17(21)23)22-18(24)11-5-3-4-6-12(11)19(22)25/h1,8-9H,3-7,10H2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=FOUWCSDKDDHKQP-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as benzoxazinones. These are organic compounds containing a benzene fused to an oxazine ring (a six-member aliphatic ring with four carbon atoms, one oxygen atom, and one nitrogen atom) bearing a ketone group. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzoxazines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Benzoxazinones | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Benzoxazinones | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | This is a man-made compound that is used as a pesticide. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB34854 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 92425 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL2133606 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 83443 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C11035 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D3847.pdf | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- This enzyme metabolizes arachidonic acid predominantly via a NADPH-dependent olefin epoxidation to all four regioisomeric cis-epoxyeicosatrienoic acids. One of the predominant enzymes responsible for the epoxidation of endogenous cardiac arachidonic acid pools.
- Gene Name:
- CYP2J2
- Uniprot ID:
- P51589
- Molecular Weight:
- 57610.165 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.50 uM | NVS_ADME_hCYP2J2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.60 uM | CLZD_CYP2B6_6 | CellzDirect |
| AC50 | 3.60 uM | CLZD_CYP2B6_48 | CellzDirect |
| AC50 | 0.60 uM | CLZD_CYP2B6_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Receptor tyrosine kinase binding ligands of the EGF family and activating several signaling cascades to convert extracellular cues into appropriate cellular responses. Known ligands include EGF, TGFA/TGF-alpha, amphiregulin, epigen/EPGN, BTC/betacellulin, epiregulin/EREG and HBEGF/heparin-binding EGF. Ligand binding triggers receptor homo- and/or heterodimerization and autophosphorylation on key cytoplasmic residues. The phosphorylated receptor recruits adapter proteins like GRB2 which in turn activates complex downstream signaling cascades. Activates at least 4 major downstream signaling cascades including the RAS-RAF-MEK-ERK, PI3 kinase-AKT, PLCgamma-PKC and STATs modules. May also activate the NF-kappa-B signaling cascade. Also directly phosphorylates other proteins like RGS16, activating its GTPase activity and probably coupling the EGF receptor signaling to the G protein-coupled receptor signaling. Also phosphorylates MUC1 and increases its interaction with SRC and CTNNB1/beta-catenin.Isoform 2 may act as an antagonist of EGF action.
- Gene Name:
- EGFR
- Uniprot ID:
- P00533
- Molecular Weight:
- 134276.185 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_EGFR_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.77 uM | CLZD_CYP3A4_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]