| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2013-04-25 07:56:52 UTC |
|---|
| Update Date | 2014-12-24 20:26:33 UTC |
|---|
| Accession Number | T3D3856 |
|---|
| Identification |
|---|
| Common Name | Halosulfuron-methyl |
|---|
| Class | Small Molecule |
|---|
| Description | Halosulfuron-methyl is a selective herbicide for post-emergence control of sedges and other weeds in turf. It is also used on maize, sugarcane and rice. It interferes with the function of the acetolactate synthase enzyme, resulting in a rapid cessation of cell division and plant growth in both roots and shoots. In sulfite-sensitive individuals, skin reactions have been reported following dermal exposure. |
|---|
| Compound Type | - Amide
- Amine
- Ester
- Ether
- Food Toxin
- Herbicide
- Household Toxin
- Metabolite
- Organic Compound
- Organochloride
- Pesticide
- Synthetic Compound
|
|---|
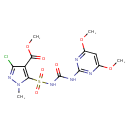
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | A 841101 | | Battalion | | C.i. Natural Brown 3 | | Halosulfuron-methyl, BSI | | Inpool | | Manage | | MON 12000 | | MON 12037 | | NC 319 | | Permit | | Permit 75WG | | Sandea | | Sempra |
|
|---|
| Chemical Formula | C13H15ClN6O7S |
|---|
| Average Molecular Mass | 434.812 g/mol |
|---|
| Monoisotopic Mass | 434.041 g/mol |
|---|
| CAS Registry Number | 100784-20-1 |
|---|
| IUPAC Name | methyl 3-chloro-5-({[(4,6-dimethoxypyrimidin-2-yl)carbamoyl]amino}sulfonyl)-1-methyl-1H-pyrazole-4-carboxylate |
|---|
| Traditional Name | methyl 3-chloro-5-{[(4,6-dimethoxypyrimidin-2-yl)carbamoyl]aminosulfonyl}-1-methylpyrazole-4-carboxylate |
|---|
| SMILES | COC(=O)C1=C(N(C)N=C1Cl)S(=O)(=O)NC(O)=NC1=NC(OC)=CC(OC)=N1 |
|---|
| InChI Identifier | InChI=1S/C13H15ClN6O7S/c1-20-10(8(9(14)18-20)11(21)27-4)28(23,24)19-13(22)17-12-15-6(25-2)5-7(16-12)26-3/h5H,1-4H3,(H2,15,16,17,19,22) |
|---|
| InChI Key | InChIKey=FMGZEUWROYGLAY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrazole carboxylic acids and derivatives. These are heterocyclic compounds containing a pyrazole ring in which a hydrogen atom is replaced by a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Azoles |
|---|
| Sub Class | Pyrazoles |
|---|
| Direct Parent | Pyrazole carboxylic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrazole-4-carboxylic acid or derivatives
- Alkyl aryl ether
- Sulfonylurea
- Aryl chloride
- Aryl halide
- Pyrimidine
- Organic sulfonic acid or derivatives
- Organosulfonic acid or derivatives
- Sulfonyl
- Heteroaromatic compound
- Aminosulfonyl compound
- Methyl ester
- Vinylogous halide
- Vinylogous amide
- Carboxylic acid ester
- Azacycle
- Ether
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carboxylic acid derivative
- Carboximidic acid derivative
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Organochloride
- Organohalogen compound
- Organonitrogen compound
- Organopnictogen compound
- Organooxygen compound
- Organosulfur compound
- Organic oxide
- Hydrocarbon derivative
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 176 - 177°C | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zgi-2910300000-aa7bd5d3d54e66da1c95 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-0140900000-df3dac23df879e904830 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0791100000-099d20b30baf19251ebe | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08mi-7911000000-e78bfbaec9d2fb4dbf63 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ue9-0721900000-674a20af190d7c5ee689 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zmi-7591200000-b4cbdeacb3a5ba2651f3 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bvi-9641000000-ce6458bea764d344b753 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0fai-0090700000-2917b709870cdfbfb655 | 2021-09-24 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ufr-1890100000-e44cf4fb4eb079c13505 | 2021-09-24 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uml-9801000000-7ca96d6a1a5093033441 | 2021-09-24 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001r-0700900000-7fccdc787261e3bf0ba0 | 2021-09-24 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-0900100000-9adce7353aa7173b56f5 | 2021-09-24 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014r-1922100000-59398c62ccafe99ebdb1 | 2021-09-24 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | Not Available | 2014-09-20 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100.40 MHz, CDCl3, experimental) | Not Available | 2014-09-23 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | This is a man-made compound that is used as a pesticide. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | HMDB34859 |
|---|
| PubChem Compound ID | 91763 |
|---|
| ChEMBL ID | CHEMBL2140532 |
|---|
| ChemSpider ID | 82861 |
|---|
| KEGG ID | C18442 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D3856.pdf |
|---|
| General References | - Yannai, Shmuel. (2004) Dictionary of food compounds with CD-ROM: Additives, flavors, and ingredients. Boca Raton: Chapman & Hall/CRC.
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|