Pyruvic acid (T3D4325)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2014-08-29 06:27:15 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:46 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D4325 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Pyruvic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Pyruvic acid is an intermediate compound in the metabolism of carbohydrates, proteins, and fats. In thiamine deficiency, its oxidation is retarded and it accumulates in the tissues, especially in nervous structures. (From Stedman, 26th ed.) Biological Source: Intermediate in primary metabolism including fermentation processes. Present in muscle in redox equilibrium with Lactic acid. A common constituent, as a chiral cyclic acetal linked to saccharide residues, of bacterial polysaccharides. Isolated from cane sugar fermentation broth and peppermint. Constituent of Bauhinia purpurea, Cicer arietinum (chickpea), Delonix regia, Pisum sativum (pea) and Trigonella caerulea (sweet trefoil) Use/Importance: Reagent for regeneration of carbonyl compdounds from semicarbazones, phenylhydrazones and oximes. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
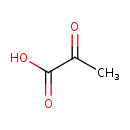
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C3H4O3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 88.062 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 88.016 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 127-17-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 2-oxopropanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | pyruvic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CC(=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C3H4O3/c1-2(4)3(5)6/h1H3,(H,5,6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=LCTONWCANYUPML-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as alpha-keto acids and derivatives. These are organic compounds containing an aldehyde substituted with a keto group on the adjacent carbon. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alpha-keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Alpha-keto acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Endogenous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Liquid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Pyruvate is absorbed from the gastrointestinal tract from whence it is transported to the liver via the portal circulation. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Pyruvate serves as a biological fuel by being converted to acetyl coenzyme A, which enters the tricarboxylic acid or Krebs cycle where it is metabolized to produce ATP aerobically. Energy can also be obtained anaerobically from pyruvate via its conversion to lactate. Pyruvate injections or perfusions increase contractile function of hearts when metabolizing glucose or fatty acids. This inotropic effect is striking in hearts stunned by ischemia/reperfusion. The inotropic effect of pyruvate requires intracoronary infusion. Among possible mechanisms for this effect are increased generation of ATP and an increase in ATP phosphorylation potential. Another is activation of pyruvate dehydrogenase, promoting its own oxidation by inhibiting pyruvate dehydrogenase kinase. Pyruvate dehydrogenase is inactivated in ischemia myocardium. Yet another is reduction of cytosolic inorganic phosphate concentration. Pyruvate, as an antioxidant, is known to scavenge such reactive oxygen species as hydrogen peroxide and lipid peroxides. Indirectly, supraphysiological levels of pyruvate may increase cellular reduced glutathione. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | In the liver, pyruvate is metabolized via several pathways. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | For nutritional supplementation, also for treating dietary shortage or imbalance | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB00119 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB00243 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 1060 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL1162144 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 1031 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C00022 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 32816 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | PYRUVATE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | PYR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Pyruvic_acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Tadamitsu Kiyoura, “Process for producing salts of pyruvic acid.” U.S. Patent US4242525, issued December, 1965. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Symporter activity
- Specific Function:
- Proton-coupled monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate. Depending on the tissue and on cicumstances, mediates the import or export of lactic acid and ketone bodies. Required for normal nutrient assimilation, increase of white adipose tissue and body weight gain when on a high-fat diet. Plays a role in cellular responses to a high-fat diet by modulating the cellular levels of lactate and pyruvate, small molecules that contribute to the regulation of central metabolic pathways and insulin secretion, with concomitant effects on plasma insulin levels and blood glucose homeostasis.
- Gene Name:
- SLC16A1
- Uniprot ID:
- P53985
- Molecular Weight:
- 53943.685 Da
References
- Duerr JM, Tucker K: Pyruvate transport in isolated cardiac mitochondria from two species of amphibian exhibiting dissimilar aerobic scope: Bufo marinus and Rana catesbeiana. J Exp Zool A Ecol Genet Physiol. 2007 Aug 1;307(8):425-38. [17583564 ]
- Han M, Trotta P, Coleman C, Linask KK: MCT-4, A511/Basigin and EF5 expression patterns during early chick cardiomyogenesis indicate cardiac cell differentiation occurs in a hypoxic environment. Dev Dyn. 2006 Jan;235(1):124-31. [16110503 ]
- Shimoyama Y, Akihara Y, Kirat D, Iwano H, Hirayama K, Kagawa Y, Ohmachi T, Matsuda K, Okamoto M, Kadosawa T, Yokota H, Taniyama H: Expression of monocarboxylate transporter 1 in oral and ocular canine melanocytic tumors. Vet Pathol. 2007 Jul;44(4):449-57. [17606506 ]
- Shimada A, Nakagawa Y, Morishige H, Yamamoto A, Fujita T: Functional characteristics of H+ -dependent nicotinate transport in primary cultures of astrocytes from rat cerebral cortex. Neurosci Lett. 2006 Jan 16;392(3):207-12. Epub 2005 Oct 5. [16213084 ]
- Philp A, Macdonald AL, Watt PW: Lactate--a signal coordinating cell and systemic function. J Exp Biol. 2005 Dec;208(Pt 24):4561-75. [16326938 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-coupled monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate. Functions as high-affinity pyruvate transporter.
- Gene Name:
- SLC16A7
- Uniprot ID:
- O60669
- Molecular Weight:
- 52199.745 Da
References
- Bonen A, Heynen M, Hatta H: Distribution of monocarboxylate transporters MCT1-MCT8 in rat tissues and human skeletal muscle. Appl Physiol Nutr Metab. 2006 Feb;31(1):31-9. [16604139 ]
- Hinoi E, Takarada T, Tsuchihashi Y, Fujimori S, Moriguchi N, Wang L, Uno K, Yoneda Y: A molecular mechanism of pyruvate protection against cytotoxicity of reactive oxygen species in osteoblasts. Mol Pharmacol. 2006 Sep;70(3):925-35. Epub 2006 Jun 9. [16766717 ]
- Yoshida Y, Holloway GP, Ljubicic V, Hatta H, Spriet LL, Hood DA, Bonen A: Negligible direct lactate oxidation in subsarcolemmal and intermyofibrillar mitochondria obtained from red and white rat skeletal muscle. J Physiol. 2007 Aug 1;582(Pt 3):1317-35. Epub 2007 Jun 7. [17556391 ]
- de Laplanche E, Gouget K, Cleris G, Dragounoff F, Demont J, Morales A, Bezin L, Godinot C, Perriere G, Mouchiroud D, Simonnet H: Physiological oxygenation status is required for fully differentiated phenotype in kidney cortex proximal tubules. Am J Physiol Renal Physiol. 2006 Oct;291(4):F750-60. Epub 2006 Apr 4. [16597615 ]
- Pierre K, Pellerin L: Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem. 2005 Jul;94(1):1-14. [15953344 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity).
- Gene Name:
- SLC16A4
- Uniprot ID:
- O15374
- Molecular Weight:
- 54021.43 Da
References
- Kay HH, Zhu S, Tsoi S: Hypoxia and lactate production in trophoblast cells. Placenta. 2007 Aug-Sep;28(8-9):854-60. Epub 2007 Feb 2. [17275903 ]
- Han M, Trotta P, Coleman C, Linask KK: MCT-4, A511/Basigin and EF5 expression patterns during early chick cardiomyogenesis indicate cardiac cell differentiation occurs in a hypoxic environment. Dev Dyn. 2006 Jan;235(1):124-31. [16110503 ]
- Bonen A, Heynen M, Hatta H: Distribution of monocarboxylate transporters MCT1-MCT8 in rat tissues and human skeletal muscle. Appl Physiol Nutr Metab. 2006 Feb;31(1):31-9. [16604139 ]
- Pierre K, Pellerin L: Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem. 2005 Jul;94(1):1-14. [15953344 ]
- Shimada A, Nakagawa Y, Morishige H, Yamamoto A, Fujita T: Functional characteristics of H+ -dependent nicotinate transport in primary cultures of astrocytes from rat cerebral cortex. Neurosci Lett. 2006 Jan 16;392(3):207-12. Epub 2005 Oct 5. [16213084 ]
- General Function:
- Pyruvate carboxylase activity
- Specific Function:
- Pyruvate carboxylase catalyzes a 2-step reaction, involving the ATP-dependent carboxylation of the covalently attached biotin in the first step and the transfer of the carboxyl group to pyruvate in the second. Catalyzes in a tissue specific manner, the initial reactions of glucose (liver, kidney) and lipid (adipose tissue, liver, brain) synthesis from pyruvate.
- Gene Name:
- PC
- Uniprot ID:
- P11498
- Molecular Weight:
- 129632.565 Da
References
- Jitrapakdee S, Vidal-Puig A, Wallace JC: Anaplerotic roles of pyruvate carboxylase in mammalian tissues. Cell Mol Life Sci. 2006 Apr;63(7-8):843-54. [16505973 ]
- Simpson NE, Khokhlova N, Oca-Cossio JA, Constantinidis I: Insights into the role of anaplerosis in insulin secretion: A 13C NMR study. Diabetologia. 2006 Jun;49(6):1338-48. Epub 2006 Mar 31. [16575559 ]
- Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB: Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J Biol Chem. 2006 Aug 4;281(31):22342-51. Epub 2006 Jun 1. [16740637 ]
- Ikeda K, Yukihiro Hiraoka B, Iwai H, Matsumoto T, Mineki R, Taka H, Takamori K, Ogawa H, Yamakura F: Detection of 6-nitrotryptophan in proteins by Western blot analysis and its application for peroxynitrite-treated PC12 cells. Nitric Oxide. 2007 Feb;16(1):18-28. Epub 2006 May 4. [16765071 ]
- Liu L, Li Y, Zhu Y, Du G, Chen J: Redistribution of carbon flux in Torulopsis glabrata by altering vitamin and calcium level. Metab Eng. 2007 Jan;9(1):21-9. Epub 2006 Aug 12. [17008113 ]
- General Function:
- Pyruvate kinase activity
- Specific Function:
- Plays a key role in glycolysis.
- Gene Name:
- PKLR
- Uniprot ID:
- P30613
- Molecular Weight:
- 61829.575 Da
References
- Percy MJ, van Wijk R, Haggan S, Savage GA, Boyd K, Dempsey S, Hamilton J, Kettle P, Kyle A, Shepherd CW, van Solinge WW, Lappin TR, McMullin MF: Pyruvate kinase deficient hemolytic anemia in the Northern Irish population. Blood Cells Mol Dis. 2007 Sep-Oct;39(2):189-94. Epub 2007 Jun 15. [17574881 ]
- Meza NW, Quintana-Bustamante O, Puyet A, Rio P, Navarro S, Diez A, Bueren JA, Bautista JM, Segovia JC: In vitro and in vivo expression of human erythrocyte pyruvate kinase in erythroid cells: a gene therapy approach. Hum Gene Ther. 2007 Jun;18(6):502-14. [17547515 ]
- Rajaseger G, Lim CL, Lee KW, Arjunan P, Jia L, Moochhala S: Profiling of hepatocellular proteins by 1D PAGE-MALDI/MS/MS in a rat heat stress model. Front Biosci. 2006 Sep 1;11:2924-8. [16720364 ]
- Xu J, Christian B, Jump DB: Regulation of rat hepatic L-pyruvate kinase promoter composition and activity by glucose, n-3 polyunsaturated fatty acids, and peroxisome proliferator-activated receptor-alpha agonist. J Biol Chem. 2006 Jul 7;281(27):18351-62. Epub 2006 Apr 27. [16644726 ]
- Suzuki T, Kawamoto M, Murai A, Muramatsu T: Identification of the regulatory region of the L-type pyruvate kinase gene in mouse liver by hydrodynamics-based gene transfection. J Nutr. 2006 Jan;136(1):16-20. [16365052 ]
- General Function:
- Pyruvate kinase activity
- Specific Function:
- Glycolytic enzyme that catalyzes the transfer of a phosphoryl group from phosphoenolpyruvate (PEP) to ADP, generating ATP. Stimulates POU5F1-mediated transcriptional activation. Plays a general role in caspase independent cell death of tumor cells. The ratio betwween the highly active tetrameric form and nearly inactive dimeric form determines whether glucose carbons are channeled to biosynthetic processes or used for glycolytic ATP production. The transition between the 2 forms contributes to the control of glycolysis and is important for tumor cell proliferation and survival.
- Gene Name:
- PKM
- Uniprot ID:
- P14618
- Molecular Weight:
- 57936.38 Da
References
- Li Y, Chang Y, Zhang L, Feng Q, Liu Z, Zhang Y, Zuo J, Meng Y, Fang F: High glucose upregulates pantothenate kinase 4 (PanK4) and thus affects M2-type pyruvate kinase (Pkm2). Mol Cell Biochem. 2005 Sep;277(1-2):117-25. [16132722 ]
- Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A: Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007 Feb 15;67(4):1602-8. [17308100 ]
- Vlaeminck-Guillem V, Safi R, Guillem P, Leteurtre E, Duterque-Coquillaud M, Laudet V: Thyroid hormone receptor expression in the obligatory paedomorphic salamander Necturus maculosus. Int J Dev Biol. 2006;50(6):553-60. [16741870 ]
- Weinberger R, Appel B, Stein A, Metz Y, Neheman A, Barak M: The pyruvate kinase isoenzyme M2 (Tu M2-PK) as a tumour marker for renal cell carcinoma. Eur J Cancer Care (Engl). 2007 Jul;16(4):333-7. [17587357 ]
- Staib P, Hoffmann M, Schinkothe T: Plasma levels of tumor M2-pyruvate kinase should not be used as a tumor marker for hematological malignancies and solid tumors. Clin Chem Lab Med. 2006;44(1):28-31. [16375581 ]
- General Function:
- Succinate-semialdehyde dehydrogenase binding
- Specific Function:
- Catalyzes the conversion of gamma-aminobutyrate and L-beta-aminoisobutyrate to succinate semialdehyde and methylmalonate semialdehyde, respectively. Can also convert delta-aminovalerate and beta-alanine.
- Gene Name:
- ABAT
- Uniprot ID:
- P80404
- Molecular Weight:
- 56438.405 Da
References
- Andersen G, Andersen B, Dobritzsch D, Schnackerz KD, Piskur J: A gene duplication led to specialized gamma-aminobutyrate and beta-alanine aminotransferase in yeast. FEBS J. 2007 Apr;274(7):1804-17. Epub 2007 Mar 12. [17355287 ]
- Schmidt C, Hofmann U, Kohlmuller D, Murdter T, Zanger UM, Schwab M, Hoffmann GF: Comprehensive analysis of pyrimidine metabolism in 450 children with unspecific neurological symptoms using high-pressure liquid chromatography-electrospray ionization tandem mass spectrometry. J Inherit Metab Dis. 2005;28(6):1109-22. [16435204 ]
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- General Function:
- Pyridoxal phosphate binding
- Specific Function:
- Can metabolize asymmetric dimethylarginine (ADMA) via transamination to alpha-keto-delta-(NN-dimethylguanidino) valeric acid (DMGV). ADMA is a potent inhibitor of nitric-oxide (NO) synthase, and this activity provides mechanism through which the kidney regulates blood pressure.
- Gene Name:
- AGXT2
- Uniprot ID:
- Q9BYV1
- Molecular Weight:
- 57155.905 Da
References
- Overington JP, Al-Lazikani B, Hopkins AL: How many drug targets are there? Nat Rev Drug Discov. 2006 Dec;5(12):993-6. [17139284 ]
- Imming P, Sinning C, Meyer A: Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006 Oct;5(10):821-34. [17016423 ]
- Tamaki N, Fujimoto S, Mizota C, Kaneko M, Kikugawa M: Inhibitory effect of 6-azauracil on beta-alanine metabolism in rat. J Nutr Sci Vitaminol (Tokyo). 1989 Oct;35(5):451-61. [2632679 ]
- General Function:
- Oxidative rna demethylase activity
- Specific Function:
- Dioxygenase that repairs alkylated DNA and RNA by oxidative demethylation. Has highest activity towards single-stranded RNA containing 3-methyluracil, followed by single-stranded DNA containing 3-methylthymine. Has low demethylase activity towards single-stranded DNA containing 1-methyladenine or 3-methylcytosine (PubMed:18775698, PubMed:20376003). Specifically demethylates N(6)-methyladenosine (m6A) RNA, the most prevalent internal modification of messenger RNA (mRNA) in higher eukaryotes (PubMed:22002720, PubMed:26458103). Has no activity towards 1-methylguanine. Has no detectable activity towards double-stranded DNA. Requires molecular oxygen, alpha-ketoglutarate and iron. Contributes to the regulation of the global metabolic rate, energy expenditure and energy homeostasis. Contributes to the regulation of body size and body fat accumulation (PubMed:18775698, PubMed:20376003).
- Gene Name:
- FTO
- Uniprot ID:
- Q9C0B1
- Molecular Weight:
- 58281.53 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >1000 uM | Not Available | BindingDB 19473 |
References
- Aik W, Demetriades M, Hamdan MK, Bagg EA, Yeoh KK, Lejeune C, Zhang Z, McDonough MA, Schofield CJ: Structural basis for inhibition of the fat mass and obesity associated protein (FTO). J Med Chem. 2013 May 9;56(9):3680-8. doi: 10.1021/jm400193d. Epub 2013 Apr 23. [23547775 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity).
- Gene Name:
- SLC16A8
- Uniprot ID:
- O95907
- Molecular Weight:
- 52318.215 Da
References
- Jansen S, Esmaeilpour T, Pantaleon M, Kaye PL: Glucose affects monocarboxylate cotransporter (MCT) 1 expression during mouse preimplantation development. Reproduction. 2006 Mar;131(3):469-79. [16514190 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity).
- Gene Name:
- SLC16A3
- Uniprot ID:
- O15427
- Molecular Weight:
- 49468.9 Da
References
- Shimada A, Nakagawa Y, Morishige H, Yamamoto A, Fujita T: Functional characteristics of H+ -dependent nicotinate transport in primary cultures of astrocytes from rat cerebral cortex. Neurosci Lett. 2006 Jan 16;392(3):207-12. Epub 2005 Oct 5. [16213084 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity).
- Gene Name:
- SLC16A5
- Uniprot ID:
- O15375
- Molecular Weight:
- 54993.04 Da
References
- Bonen A, Heynen M, Hatta H: Distribution of monocarboxylate transporters MCT1-MCT8 in rat tissues and human skeletal muscle. Appl Physiol Nutr Metab. 2006 Feb;31(1):31-9. [16604139 ]
- General Function:
- Symporter activity
- Specific Function:
- Proton-linked monocarboxylate transporter. Catalyzes the rapid transport across the plasma membrane of many monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and the ketone bodies acetoacetate, beta-hydroxybutyrate and acetate (By similarity).
- Gene Name:
- SLC16A6
- Uniprot ID:
- O15403
- Molecular Weight:
- 57392.83 Da
References
- Bonen A, Heynen M, Hatta H: Distribution of monocarboxylate transporters MCT1-MCT8 in rat tissues and human skeletal muscle. Appl Physiol Nutr Metab. 2006 Feb;31(1):31-9. [16604139 ]
- General Function:
- Transporter activity
- Specific Function:
- Very active and specific thyroid hormone transporter. Stimulates cellular uptake of thyroxine (T4), triiodothyronine (T3), reverse triiodothyronine (rT3) and diidothyronine. Does not transport Leu, Phe, Trp or Tyr.
- Gene Name:
- SLC16A2
- Uniprot ID:
- P36021
- Molecular Weight:
- 59510.86 Da
References
- Bonen A, Heynen M, Hatta H: Distribution of monocarboxylate transporters MCT1-MCT8 in rat tissues and human skeletal muscle. Appl Physiol Nutr Metab. 2006 Feb;31(1):31-9. [16604139 ]
- General Function:
- Pyruvate dehydrogenase activity
- Specific Function:
- The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2), and thereby links the glycolytic pathway to the tricarboxylic cycle.
- Gene Name:
- PDHB
- Uniprot ID:
- P11177
- Molecular Weight:
- 39233.1 Da
References
- Kumar V, Rangaraj N, Shivaji S: Activity of pyruvate dehydrogenase A (PDHA) in hamster spermatozoa correlates positively with hyperactivation and is associated with sperm capacitation. Biol Reprod. 2006 Nov;75(5):767-77. Epub 2006 Jul 19. [16855207 ]
- General Function:
- Sodium-independent organic anion transmembrane transporter activity
- Specific Function:
- May mediate the release of newly synthesized prostaglandins from cells, the transepithelial transport of prostaglandins, and the clearance of prostaglandins from the circulation. Transports PGD2, as well as PGE1, PGE2 and PGF2A.
- Gene Name:
- SLCO2A1
- Uniprot ID:
- Q92959
- Molecular Weight:
- 70043.33 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 26 uM | Not Available | BindingDB 19473 |
References
- Chan BS, Endo S, Kanai N, Schuster VL: Identification of lactate as a driving force for prostanoid transport by prostaglandin transporter PGT. Am J Physiol Renal Physiol. 2002 Jun;282(6):F1097-102. [11997326 ]