| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-09-11 05:16:26 UTC |
|---|
| Update Date | 2014-12-24 20:26:57 UTC |
|---|
| Accession Number | T3D4784 |
|---|
| Identification |
|---|
| Common Name | Clofibrate |
|---|
| Class | Small Molecule |
|---|
| Description | A fibric acid derivative used in the treatment of hyperlipoproteinemia type III and severe hypertriglyceridemia. |
|---|
| Compound Type | - Anticholesteremic Agent
- Drug
- Ester
- Ether
- Hypolipidemic Agent
- Metabolite
- Organic Compound
- Organochloride
- Synthetic Compound
|
|---|
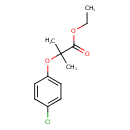
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | 2-(4-Chlorophenoxy)-2-methylpropanoic acid ethyl ester | | 2-(p-Chlorophenoxy)-2-methylpropionic acid ethyl ester | | alpha-(p-Chlorophenoxy)isobutyric acid, ethyl ester | | alpha-p-Chlorophenoxyisobutyryl ethyl ester | | Alufibrate | | Atromid-S | | Binograc | | Chlorfenisate | | Chlorphenisate | | Clobrate | | Clofibate | | Clofibrato | | Clofibratum | | Clofibric acid | | CPIB | | ELPI | | EPIB | | Ethyl 2-(P-chlorophenoxy)isobutyrate | | Ethyl chlorophenoxyisobutyrate | | Ethyl clofibrate | | Ethyl p-chlorophenoxyisobutyrate | | Ethyl para-chlorophenoxyisobutyrate | | Hisunsero | | Koliva | | Lipofacton | | Liprin | | Myanlin |
|
|---|
| Chemical Formula | C12H15ClO3 |
|---|
| Average Molecular Mass | 242.699 g/mol |
|---|
| Monoisotopic Mass | 242.071 g/mol |
|---|
| CAS Registry Number | 637-07-0 |
|---|
| IUPAC Name | ethyl 2-(4-chlorophenoxy)-2-methylpropanoate |
|---|
| Traditional Name | artes |
|---|
| SMILES | CCOC(=O)C(C)(C)OC1=CC=C(Cl)C=C1 |
|---|
| InChI Identifier | InChI=1S/C12H15ClO3/c1-4-15-11(14)12(2,3)16-10-7-5-9(13)6-8-10/h5-8H,4H2,1-3H3 |
|---|
| InChI Key | InChIKey=KNHUKKLJHYUCFP-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenoxyacetic acid derivatives. Phenoxyacetic acid derivatives are compounds containing an anisole where the methane group is linked to an acetic acid or a derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Phenoxyacetic acid derivatives |
|---|
| Direct Parent | Phenoxyacetic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenoxyacetate
- Phenoxy compound
- Phenol ether
- Alkyl aryl ether
- Chlorobenzene
- Halobenzene
- Aryl chloride
- Aryl halide
- Carboxylic acid ester
- Carboxylic acid derivative
- Ether
- Monocarboxylic acid or derivatives
- Organochloride
- Organooxygen compound
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organohalogen compound
- Organic oxide
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | 118-119°C | | Boiling Point | 149°C at 2.00E+01 mm Hg | | Solubility | Insoluble | | LogP | 3.3 |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00or-4900000000-b0f27a4473b7e59d68c3 | 2017-09-01 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-014v-0930000000-829386905d164431d8d2 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-014r-0900000000-7d85ceb3a8f1381c7da6 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001l-0900000000-4a173dddc088d2ecd596 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-015c-0900000000-dcd68fabfcd01b516107 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-014r-0900000000-409b5800e94ffbb11863 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001l-0900000000-63251a4c6680c9389c2f | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0290000000-f8e44478c85e8deb28e7 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004l-1960000000-5686cbdf03cff625cebc | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-2900000000-de1ce5552c1bea344c1e | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0920000000-255926cbf0927bf8a9fa | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-4910000000-64c9da59c5fb22afb366 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0059-7900000000-cd5c6706685e8cd7dbe1 | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0390000000-a206c9f16b476083dcdb | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1920000000-cdf7fd63fdcb1f2840cd | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004i-6900000000-bf1eef6ec7349495a9b5 | 2016-08-03 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-6980000000-60ef91c08059dec83cfe | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00p0-7910000000-106c9afa2a025a09891e | 2021-10-11 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9200000000-710d7a203fef2cd8e30b | 2021-10-11 | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-004i-3900000000-fc2f01df51d424557e9f | 2014-09-20 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Completely but slowly absorbed from the intestine. Between 95% and 99% of an oral dose of clofibrate is excreted in the urine as free and conjugated clofibric acid; thus, the absorption of clofibrate is virtually complete. |
|---|
| Mechanism of Toxicity | Clofibrate increases the activity of extrahepatic lipoprotein lipase (LL), thereby increasing lipoprotein triglyceride lipolysis. Chylomicrons are degraded, VLDLs are converted to LDLs, and LDLs are converted to HDL. This is accompanied by a slight increase in secretion of lipids into the bile and ultimately the intestine. Clofibrate also inhibits the synthesis and increases the clearance of apolipoprotein B, a carrier molecule for VLDL. Also, as a fibrate, Clofibrate is an agonist of the PPAR-‘± receptor[4] in muscle, liver, and other tissues. This agonism ultimately leads to modification in gene expression resulting in increased beta-oxidation, decreased triglyceride secretion, increased HDL, increased lipoprotein lipase activity. |
|---|
| Metabolism | Hepatic and gastrointestinal: rapid de-esterification occurs in the gastrointestinal tract and/or on first-pass metabolism to produce the active form, clofibric acid (chlorophenoxy isobutyric acid [CPIB]).
Half Life: Half-life in normal volunteers averages 18 to 22 hours (range 14 to 35 hours) but can vary by up to 7 hours in the same subject at different times. |
|---|
| Toxicity Values | Oral, mouse: LD50 = 1220 mg/kg; Oral, rabbit: LD50 = 1370 mg/kg; Oral, rat: LD50 = 940 mg/kg. |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | 3, not classifiable as to its carcinogenicity to humans. (1) |
|---|
| Uses/Sources | For Primary Dysbetalipoproteinemia (Type III hyperlipidemia) that does not respond adequately to diet. This helps control high cholesterol and high triglyceride levels. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB00636 |
|---|
| HMDB ID | HMDB14774 |
|---|
| PubChem Compound ID | 2796 |
|---|
| ChEMBL ID | CHEMBL565 |
|---|
| ChemSpider ID | 2694 |
|---|
| KEGG ID | C06916 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 3750 |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Clofibrate |
|---|
| References |
|---|
| Synthesis Reference | Jones, W.G.M.,Thorp, J.M. and Waring, W.S.; U.S. Patent 3,262,850; July 26, 1966; assigned to Imperial Chemical Industries Limited, England. |
|---|
| MSDS | Link |
|---|
| General References | - International Agency for Research on Cancer (2014). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Link]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|