You are using an unsupported browser. Please upgrade your browser to a newer version to get the best experience on Toxin, Toxin Target Database.
Astemizole (T3D2844)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-07-21 20:27:18 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:25:52 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D2844 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Astemizole | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Astemizole is a long-acting, non-sedating second generation antihistamine used in the treatment of allergy symptoms. It was withdrawn from market by the manufacturer in 1999 due to the potential to cause arrhythmias at high doses, especially when when taken with CYP inhibitors or grapefruit juice. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
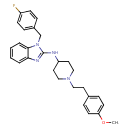
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C28H31FN4O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 458.570 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 458.248 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 68844-77-9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-1,3-benzodiazol-2-amine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | astemizole | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | COC1=CC=C(CCN2CCC(CC2)N=C2NC3=CC=CC=C3N2CC2=CC=C(F)C=C2)C=C1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C28H31FN4O/c1-34-25-12-8-21(9-13-25)14-17-32-18-15-24(16-19-32)30-28-31-26-4-2-3-5-27(26)33(28)20-22-6-10-23(29)11-7-22/h2-13,24H,14-20H2,1H3,(H,30,31) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=GXDALQBWZGODGZ-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as benzimidazoles. These are organic compounds containing a benzene ring fused to an imidazole ring (five member ring containing a nitrogen atom, 4 carbon atoms, and two double bonds). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzimidazoles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Benzimidazoles | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Oral. Rapidly absorbed from the gastrointestinal tract. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Astemizole competes with histamine for binding at H1-receptor sites in the GI tract, uterus, large blood vessels, and bronchial muscle. This reversible binding of astemizole to H1-receptors suppresses the formation of edema, flare, and pruritus resulting from histaminic activity. As the drug does not readily cross the blood-brain barrier and preferentially binds at H1 receptors in the peripehery rather than within the brain, CNS depression is minimal. Astemizole may also act on H3-receptors, producing adverse effects. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | It is metabolized by CYP3A4. [Wikidpedia]. Almost completely metabolized in the liver and primarily excreted in the feces. Half Life: 1 day | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 2052mg/kg (Mouse) (3) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Astemizole tablets are indicated for the relief of symptoms associated with seasonal allergic rhinitis and chronic idiopathic urticaria. (5). Astemizole was indicated for use in the relieving allergy symptoms, particularly rhinitis and conjunctivitis. It has been withdrawn from the market however due to concerns of arrhythmias. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Cardiovascular adverse events. In some cases, recognition of severe arrhythmias has been preceded by episodes of syncope. Similarly, rare cases of hypotension, palpitations, and dizziness have also been reported with Astemizole use, which may reflect undetected ventricular arrhythmia. (5) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | In the event of overdosage, supportive measures including gastric lavage and emesis should be employed. (6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | DB00637 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB14775 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 2247 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL296419 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 2160 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C06832 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 2896 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Astemizole | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Astemizole | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Godelieve Irma Christine Maria Heylen, Cornelus Gerardus Maria Janssen, Jurzak Mirek, Henricus Petrus Martinus Maria Van Assouw, “Radiolabeled astemizole and method of making.” U.S. Patent US07541476, issued June 02, 2009. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization
- Specific Function:
- Pore-forming (alpha) subunit of voltage-gated inwardly rectifying potassium channel. Channel properties are modulated by cAMP and subunit assembly. Mediates the rapidly activating component of the delayed rectifying potassium current in heart (IKr). Isoforms USO have no channel activity by themself, but modulates channel characteristics by forming heterotetramers with other isoforms which are retained intracellularly and undergo ubiquitin-dependent degradation.
- Gene Name:
- KCNH2
- Uniprot ID:
- Q12809
- Molecular Weight:
- 126653.52 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.0029 uM | Not Available | BindingDB 24226 |
| Inhibitory | 0.003 uM | Not Available | BindingDB 24226 |
| Inhibitory | 0.0128 uM | Not Available | BindingDB 24226 |
| IC50 | 0.0009 uM | Not Available | BindingDB 24226 |
| IC50 | 0.00091 uM | Not Available | BindingDB 24226 |
| IC50 | 0.000912 uM | Not Available | BindingDB 24226 |
| IC50 | 0.0015 uM | Not Available | BindingDB 24226 |
| IC50 | 0.006 uM | Not Available | BindingDB 24226 |
| IC50 | 0.008 uM | Not Available | BindingDB 24226 |
| IC50 | 0.01 uM | Not Available | BindingDB 24226 |
References
- Zhou Z, Vorperian VR, Gong Q, Zhang S, January CT: Block of HERG potassium channels by the antihistamine astemizole and its metabolites desmethylastemizole and norastemizole. J Cardiovasc Electrophysiol. 1999 Jun;10(6):836-43. [10376921 ]
- Chachin M, Katayama Y, Yamada M, Horio Y, Ohmura T, Kitagawa H, Uchida S, Kurachi Y: Epinastine, a nonsedating histamine H1 receptor antagonist, has a negligible effect on HERG channel. Eur J Pharmacol. 1999 Jun 25;374(3):457-60. [10422790 ]
- Taglialatela M, Castaldo P, Pannaccione A, Giorgio G, Genovese A, Marone G, Annunziato L: Cardiac ion channels and antihistamines: possible mechanisms of cardiotoxicity. Clin Exp Allergy. 1999 Jul;29 Suppl 3:182-9. [10444235 ]
- Grzelewska-Rzymowska I, Pietrzkowicz M, Gorska M: [The effect of second generation histamine antagonists on the heart]. Pneumonol Alergol Pol. 2001;69(3-4):217-26. [11575008 ]
- Chiu PJ, Marcoe KF, Bounds SE, Lin CH, Feng JJ, Lin A, Cheng FC, Crumb WJ, Mitchell R: Validation of a [3H]astemizole binding assay in HEK293 cells expressing HERG K+ channels. J Pharmacol Sci. 2004 Jul;95(3):311-9. [15272206 ]
- Cavalli A, Poluzzi E, De Ponti F, Recanatini M: Toward a pharmacophore for drugs inducing the long QT syndrome: insights from a CoMFA study of HERG K(+) channel blockers. J Med Chem. 2002 Aug 29;45(18):3844-53. [12190308 ]
- Pearlstein R, Vaz R, Rampe D: Understanding the structure-activity relationship of the human ether-a-go-go-related gene cardiac K+ channel. A model for bad behavior. J Med Chem. 2003 May 22;46(11):2017-22. [12747773 ]
- Becker OM, Dhanoa DS, Marantz Y, Chen D, Shacham S, Cheruku S, Heifetz A, Mohanty P, Fichman M, Sharadendu A, Nudelman R, Kauffman M, Noiman S: An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J Med Chem. 2006 Jun 1;49(11):3116-35. [16722631 ]
- Rajamani R, Tounge BA, Li J, Reynolds CH: A two-state homology model of the hERG K+ channel: application to ligand binding. Bioorg Med Chem Lett. 2005 Mar 15;15(6):1737-41. [15745831 ]
- Ermondi G, Visentin S, Caron G: GRIND-based 3D-QSAR and CoMFA to investigate topics dominated by hydrophobic interactions: the case of hERG K+ channel blockers. Eur J Med Chem. 2009 May;44(5):1926-32. doi: 10.1016/j.ejmech.2008.11.009. Epub 2008 Nov 28. [19110341 ]
- Du LP, Tsai KC, Li MY, You QD, Xia L: The pharmacophore hypotheses of I(Kr) potassium channel blockers: novel class III antiarrhythmic agents. Bioorg Med Chem Lett. 2004 Sep 20;14(18):4771-7. [15324906 ]
- Keseru GM: Prediction of hERG potassium channel affinity by traditional and hologram qSAR methods. Bioorg Med Chem Lett. 2003 Aug 18;13(16):2773-5. [12873512 ]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]
- Tobita M, Nishikawa T, Nagashima R: A discriminant model constructed by the support vector machine method for HERG potassium channel inhibitors. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2886-90. [15911273 ]
- Jia L, Sun H: Support vector machines classification of hERG liabilities based on atom types. Bioorg Med Chem. 2008 Jun 1;16(11):6252-60. doi: 10.1016/j.bmc.2008.04.028. Epub 2008 Apr 16. [18448342 ]
- Singleton DH, Boyd H, Steidl-Nichols JV, Deacon M, Groot MJ, Price D, Nettleton DO, Wallace NK, Troutman MD, Williams C, Boyd JG: Fluorescently labeled analogues of dofetilide as high-affinity fluorescence polarization ligands for the human ether-a-go-go-related gene (hERG) channel. J Med Chem. 2007 Jun 28;50(13):2931-41. Epub 2007 May 31. [17536794 ]
- Taglialatela M, Secondo A, Fresi A, Rosati B, Pannaccione A, Castaldo P, Giorgio G, Wanke E, Annunziato L: Inhibition of depolarization-induced [3H]noradrenaline release from SH-SY5Y human neuroblastoma cells by some second-generation H(1) receptor antagonists through blockade of store-operated Ca(2+) channels (SOCs). Biochem Pharmacol. 2001 Nov 1;62(9):1229-38. [11705456 ]
- Wang J, Della Penna K, Wang H, Karczewski J, Connolly TM, Koblan KS, Bennett PB, Salata JJ: Functional and pharmacological properties of canine ERG potassium channels. Am J Physiol Heart Circ Physiol. 2003 Jan;284(1):H256-67. Epub 2002 Oct 3. [12388285 ]
- Taglialatela M, Pannaccione A, Castaldo P, Giorgio G, Zhou Z, January CT, Genovese A, Marone G, Annunziato L: Molecular basis for the lack of HERG K+ channel block-related cardiotoxicity by the H1 receptor blocker cetirizine compared with other second-generation antihistamines. Mol Pharmacol. 1998 Jul;54(1):113-21. [9658196 ]
- General Function:
- Histamine receptor activity
- Specific Function:
- In peripheral tissues, the H1 subclass of histamine receptors mediates the contraction of smooth muscles, increase in capillary permeability due to contraction of terminal venules, and catecholamine release from adrenal medulla, as well as mediating neurotransmission in the central nervous system.
- Gene Name:
- HRH1
- Uniprot ID:
- P35367
- Molecular Weight:
- 55783.61 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 0.0008 uM | Not Available | BindingDB 24226 |
| Inhibitory | 0.00209 uM | Not Available | BindingDB 24226 |
References
- Chen X, Ji ZL, Chen YZ: TTD: Therapeutic Target Database. Nucleic Acids Res. 2002 Jan 1;30(1):412-5. [11752352 ]
- Cavero I, Mestre M, Guillon JM, Heuillet E, Roach AG: Preclinical in vitro cardiac electrophysiology: a method of predicting arrhythmogenic potential of antihistamines in humans? Drug Saf. 1999;21 Suppl 1:19-31; discussion 81-7. [10597865 ]
- Salata JJ, Jurkiewicz NK, Wallace AA, Stupienski RF 3rd, Guinosso PJ Jr, Lynch JJ Jr: Cardiac electrophysiological actions of the histamine H1-receptor antagonists astemizole and terfenadine compared with chlorpheniramine and pyrilamine. Circ Res. 1995 Jan;76(1):110-9. [8001268 ]
- Howarth PH, Emanuel MB, Holgate ST: Astemizole, a potent histamine H1-receptor antagonist: effect in allergic rhinoconjunctivitis, on antigen and histamine induced skin weal responses and relationship to serum levels. Br J Clin Pharmacol. 1984 Jul;18(1):1-8. [6146346 ]
- Kaliner MA, Check WA: Non-sedating antihistamines. Allergy Proc. 1988 Nov-Dec;9(6):649-63. [3147222 ]
- Llenas J, Cardelus I, Heredia A, de Mora F, Gristwood RW: Cardiotoxicity of histamine and the possible role of histamine in the arrhythmogenesis produced by certain antihistamines. Drug Saf. 1999;21 Suppl 1:33-8; discussion 81-7. [10597866 ]
- Schotte A, Janssen PF, Gommeren W, Luyten WH, Van Gompel P, Lesage AS, De Loore K, Leysen JE: Risperidone compared with new and reference antipsychotic drugs: in vitro and in vivo receptor binding. Psychopharmacology (Berl). 1996 Mar;124(1-2):57-73. [8935801 ]
- Wagner E, Wittmann HJ, Elz S, Strasser A: Mepyramine-JNJ7777120-hybrid compounds show high affinity to hH(1)R, but low affinity to hH(4)R. Bioorg Med Chem Lett. 2011 Nov 1;21(21):6274-80. doi: 10.1016/j.bmcl.2011.09.001. Epub 2011 Sep 8. [21944853 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
References
- Nicolas JM, Whomsley R, Collart P, Roba J: In vitro inhibition of human liver drug metabolizing enzymes by second generation antihistamines. Chem Biol Interact. 1999 Nov 15;123(1):63-79. [10597902 ]
- Matsumoto S, Yamazoe Y: Involvement of multiple human cytochromes P450 in the liver microsomal metabolism of astemizole and a comparison with terfenadine. Br J Clin Pharmacol. 2001 Feb;51(2):133-42. [11259984 ]
- Cvetkovic RS, Goa KL: Lopinavir/ritonavir: a review of its use in the management of HIV infection. Drugs. 2003;63(8):769-802. [12662125 ]
- Goto A, Adachi Y, Inaba A, Nakajima H, Kobayashi H, Sakai K: Identification of human p450 isoforms involved in the metabolism of the antiallergic drug, oxatomide, and its inhibitory effect on enzyme activity. Biol Pharm Bull. 2004 May;27(5):684-90. [15133245 ]
- Goto A, Ueda K, Inaba A, Nakajima H, Kobayashi H, Sakai K: Identification of human P450 isoforms involved in the metabolism of the antiallergic drug, oxatomide, and its kinetic parameters and inhibition constants. Biol Pharm Bull. 2005 Feb;28(2):328-34. [15684493 ]
- General Function:
- Somatostatin receptor activity
- Specific Function:
- Receptor for somatostatin 28 and to a lesser extent for somatostatin-14. The activity of this receptor is mediated by G proteins which inhibit adenylyl cyclase. Increases cell growth inhibition activity of SSTR2 following heterodimerization.
- Gene Name:
- SSTR5
- Uniprot ID:
- P35346
- Molecular Weight:
- 39201.925 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 4.07 uM | Not Available | BindingDB 24226 |
References
- Martin RE, Green LG, Guba W, Kratochwil N, Christ A: Discovery of the first nonpeptidic, small-molecule, highly selective somatostatin receptor subtype 5 antagonists: a chemogenomics approach. J Med Chem. 2007 Dec 13;50(25):6291-4. Epub 2007 Nov 19. [18020390 ]
- Martin RE, Mohr P, Maerki HP, Guba W, Kuratli C, Gavelle O, Binggeli A, Bendels S, Alvarez-Sanchez R, Alker A, Polonchuk L, Christ AD: Benzoxazole piperidines as selective and potent somatostatin receptor subtype 5 antagonists. Bioorg Med Chem Lett. 2009 Nov 1;19(21):6106-13. doi: 10.1016/j.bmcl.2009.09.024. Epub 2009 Sep 12. [19786348 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 3.3 uM | Not Available | BindingDB 24226 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Syndecan binding
- Specific Function:
- Endoglycosidase that cleaves heparan sulfate proteoglycans (HSPGs) into heparan sulfate side chains and core proteoglycans. Participates in extracellular matrix (ECM) degradation and remodeling. Selectively cleaves the linkage between a glucuronic acid unit and an N-sulfo glucosamine unit carrying either a 3-O-sulfo or a 6-O-sulfo group. Can also cleave the linkage between a glucuronic acid unit and an N-sulfo glucosamine unit carrying a 2-O-sulfo group, but not linkages between a glucuronic acid unit and a 2-O-sulfated iduronic acid moiety. It is essentially inactive at neutral pH but becomes active under acidic conditions such as during tumor invasion and in inflammatory processes. Facilitates cell migration associated with metastasis, wound healing and inflammation. Enhances shedding of syndecans, and increases endothelial invasion and angiogenesis in myelomas. Acts as procoagulant by increasing the generation of activation factor X in the presence of tissue factor and activation factor VII. Increases cell adhesion to the extacellular matrix (ECM), independent of its enzymatic activity. Induces AKT1/PKB phosphorylation via lipid rafts increasing cell mobility and invasion. Heparin increases this AKT1/PKB activation. Regulates osteogenesis. Enhances angiogenesis through up-regulation of SRC-mediated activation of VEGF. Implicated in hair follicle inner root sheath differentiation and hair homeostasis.
- Gene Name:
- HPSE
- Uniprot ID:
- Q9Y251
- Molecular Weight:
- 61148.17 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Dissociation | >1000 uM | Not Available | BindingDB 24226 |
References
- Gozalbes R, Mosulen S, Orti L, Rodriguez-Diaz J, Carbajo RJ, Melnyk P, Pineda-Lucena A: Hit identification of novel heparanase inhibitors by structure- and ligand-based approaches. Bioorg Med Chem. 2013 Apr 1;21(7):1944-51. doi: 10.1016/j.bmc.2013.01.033. Epub 2013 Jan 31. [23415087 ]
- General Function:
- Histamine receptor activity
- Specific Function:
- The H4 subclass of histamine receptors could mediate the histamine signals in peripheral tissues. Displays a significant level of constitutive activity (spontaneous activity in the absence of agonist).
- Gene Name:
- HRH4
- Uniprot ID:
- Q9H3N8
- Molecular Weight:
- 44495.375 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | 7.94328 uM | Not Available | BindingDB 24226 |
References
- Wagner E, Wittmann HJ, Elz S, Strasser A: Mepyramine-JNJ7777120-hybrid compounds show high affinity to hH(1)R, but low affinity to hH(4)R. Bioorg Med Chem Lett. 2011 Nov 1;21(21):6274-80. doi: 10.1016/j.bmcl.2011.09.001. Epub 2011 Sep 8. [21944853 ]
- General Function:
- Structural constituent of cytoskeleton
- Specific Function:
- Promotes microtubule assembly and stability, and might be involved in the establishment and maintenance of neuronal polarity. The C-terminus binds axonal microtubules while the N-terminus binds neural plasma membrane components, suggesting that tau functions as a linker protein between both. Axonal polarity is predetermined by TAU/MAPT localization (in the neuronal cell) in the domain of the cell body defined by the centrosome. The short isoforms allow plasticity of the cytoskeleton whereas the longer isoforms may preferentially play a role in its stabilization.
- Gene Name:
- MAPT
- Uniprot ID:
- P10636
- Molecular Weight:
- 78927.025 Da
References
- Rojo LE, Alzate-Morales J, Saavedra IN, Davies P, Maccioni RB: Selective interaction of lansoprazole and astemizole with tau polymers: potential new clinical use in diagnosis of Alzheimer's disease. J Alzheimers Dis. 2010;19(2):573-89. doi: 10.3233/JAD-2010-1262. [20110603 ]
- General Function:
- Monovalent cation:proton antiporter activity
- Specific Function:
- Solute transporter for tetraethylammonium (TEA), 1-methyl-4-phenylpyridinium (MPP), cimetidine, N-methylnicotinamide (NMN), metformin, creatinine, guanidine, procainamide, topotecan, estrone sulfate, acyclovir, ganciclovir and also the zwitterionic cephalosporin, cephalexin and cephradin. Seems to also play a role in the uptake of oxaliplatin (a new platinum anticancer agent). Able to transport paraquat (PQ or N,N-dimethyl-4-4'-bipiridinium); a widely used herbicid. Responsible for the secretion of cationic drugs across the brush border membranes.
- Gene Name:
- SLC47A1
- Uniprot ID:
- Q96FL8
- Molecular Weight:
- 61921.585 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 26.4 uM | Not Available | BindingDB 24226 |
References
- Wittwer MB, Zur AA, Khuri N, Kido Y, Kosaka A, Zhang X, Morrissey KM, Sali A, Huang Y, Giacomini KM: Discovery of potent, selective multidrug and toxin extrusion transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug profiling and computational modeling. J Med Chem. 2013 Feb 14;56(3):781-95. doi: 10.1021/jm301302s. Epub 2013 Jan 22. [23241029 ]
- General Function:
- Xenobiotic-transporting atpase activity
- Specific Function:
- Energy-dependent efflux pump responsible for decreased drug accumulation in multidrug-resistant cells.
- Gene Name:
- ABCB1
- Uniprot ID:
- P08183
- Molecular Weight:
- 141477.255 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | 0.3 uM | Not Available | BindingDB 24226 |
| IC50 | 1.3 uM | Not Available | BindingDB 24226 |
References
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J: Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003 Apr 24;46(9):1716-25. [12699389 ]
- General Function:
- Phosphorelay sensor kinase activity
- Specific Function:
- Pore-forming (alpha) subunit of a voltage-gated delayed rectifier potassium channel (PubMed:22732247). Channel properties may be modulated by subunit assembly, but not by cyclic nucleotides (By similarity). Mediates IK(NI) current in myoblasts (PubMed:9738473). Involved in the regulation of cell proliferation and differentiation, in particular adipogenic and osteogenic differentiation in bone marrow-derived mesenchymal stem cells (MSCs) (PubMed:23881642).
- Gene Name:
- KCNH1
- Uniprot ID:
- O95259
- Molecular Weight:
- 111421.76 Da
References
- Roy J, Vantol B, Cowley EA, Blay J, Linsdell P: Pharmacological separation of hEAG and hERG K+ channel function in the human mammary carcinoma cell line MCF-7. Oncol Rep. 2008 Jun;19(6):1511-6. [18497958 ]
- General Function:
- Somatostatin receptor activity
- Specific Function:
- Receptor for somatostatin-14 and -28. This receptor is coupled via pertussis toxin sensitive G proteins to inhibition of adenylyl cyclase. In addition it stimulates phosphotyrosine phosphatase and PLC via pertussis toxin insensitive as well as sensitive G proteins. Inhibits calcium entry by suppressing voltage-dependent calcium channels. Acts as the functionally dominant somatostatin receptor in pancreatic alpha- and beta-cells where it mediates the inhibitory effect of somatostatin-14 on hormone secretion. Inhibits cell growth through enhancement of MAPK1 and MAPK2 phosphorylation and subsequent up-regulation of CDKN1B. Stimulates neuronal migration and axon outgrowth and may participate in neuron development and maturation during brain development. Mediates negative regulation of insulin receptor signaling through PTPN6. Inactivates SSTR3 receptor function following heterodimerization.
- Gene Name:
- SSTR2
- Uniprot ID:
- P30874
- Molecular Weight:
- 41332.37 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | >10 uM | Not Available | BindingDB 24226 |
References
- Martin RE, Mohr P, Maerki HP, Guba W, Kuratli C, Gavelle O, Binggeli A, Bendels S, Alvarez-Sanchez R, Alker A, Polonchuk L, Christ AD: Benzoxazole piperidines as selective and potent somatostatin receptor subtype 5 antagonists. Bioorg Med Chem Lett. 2009 Nov 1;19(21):6106-13. doi: 10.1016/j.bmcl.2009.09.024. Epub 2009 Sep 12. [19786348 ]
- General Function:
- Somatostatin receptor activity
- Specific Function:
- Receptor for somatostatin-14 and -28. This receptor is coupled via pertussis toxin sensitive G proteins to inhibition of adenylyl cyclase.
- Gene Name:
- SSTR3
- Uniprot ID:
- P32745
- Molecular Weight:
- 45846.995 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | >10 uM | Not Available | BindingDB 24226 |
References
- Martin RE, Mohr P, Maerki HP, Guba W, Kuratli C, Gavelle O, Binggeli A, Bendels S, Alvarez-Sanchez R, Alker A, Polonchuk L, Christ AD: Benzoxazole piperidines as selective and potent somatostatin receptor subtype 5 antagonists. Bioorg Med Chem Lett. 2009 Nov 1;19(21):6106-13. doi: 10.1016/j.bmcl.2009.09.024. Epub 2009 Sep 12. [19786348 ]
- General Function:
- Somatostatin receptor activity
- Specific Function:
- Receptor for somatostatin-14. The activity of this receptor is mediated by G proteins which inhibits adenylyl cyclase. It is functionally coupled not only to inhibition of adenylate cyclase, but also to activation of both arachidonate release and mitogen-activated protein (MAP) kinase cascade. Mediates antiproliferative action of somatostatin in tumor cells.
- Gene Name:
- SSTR4
- Uniprot ID:
- P31391
- Molecular Weight:
- 42002.245 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| Inhibitory | >10 uM | Not Available | BindingDB 24226 |
References
- Martin RE, Mohr P, Maerki HP, Guba W, Kuratli C, Gavelle O, Binggeli A, Bendels S, Alvarez-Sanchez R, Alker A, Polonchuk L, Christ AD: Benzoxazole piperidines as selective and potent somatostatin receptor subtype 5 antagonists. Bioorg Med Chem Lett. 2009 Nov 1;19(21):6106-13. doi: 10.1016/j.bmcl.2009.09.024. Epub 2009 Sep 12. [19786348 ]
- General Function:
- Ubiquitin-protein transferase activity
- Specific Function:
- The UBE2V1-UBE2N and UBE2V2-UBE2N heterodimers catalyze the synthesis of non-canonical 'Lys-63'-linked polyubiquitin chains. This type of polyubiquitination does not lead to protein degradation by the proteasome. Mediates transcriptional activation of target genes. Plays a role in the control of progress through the cell cycle and differentiation. Plays a role in the error-free DNA repair pathway and contributes to the survival of cells after DNA damage. Acts together with the E3 ligases, HLTF and SHPRH, in the 'Lys-63'-linked poly-ubiquitination of PCNA upon genotoxic stress, which is required for DNA repair. Appears to act together with E3 ligase RNF5 in the 'Lys-63'-linked polyubiquitination of JKAMP thereby regulating JKAMP function by decreasing its association with components of the proteasome and ERAD. Promotes TRIM5 capsid-specific restriction activity and the UBE2V1-UBE2N heterodimer acts in concert with TRIM5 to generate 'Lys-63'-linked polyubiquitin chains which activate the MAP3K7/TAK1 complex which in turn results in the induction and expression of NF-kappa-B and MAPK-responsive inflammatory genes.
- Gene Name:
- UBE2N
- Uniprot ID:
- P61088
- Molecular Weight:
- 17137.625 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| IC50 | >20 uM | Not Available | BindingDB 24226 |
References
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK: BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007 Jan;35(Database issue):D198-201. Epub 2006 Dec 1. [17145705 ]