| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2013-04-25 07:56:54 UTC |
|---|
| Update Date | 2014-12-24 20:26:34 UTC |
|---|
| Accession Number | T3D3911 |
|---|
| Identification |
|---|
| Common Name | Propoxycarbazone-sodium |
|---|
| Class | Small Molecule |
|---|
| Description | Propoxycarbazone-sodium is a postemergence herbicide used primarily to control various annual and perennial grasses in rye, triticale, and wheat. Its efficacy is the result of the inhibition of the enzyme acetolactate synthase (ALS) in target plants. |
|---|
| Compound Type | - Amide
- Ester
- Ether
- Herbicide
- Organic Compound
- Pesticide
- Synthetic Compound
|
|---|
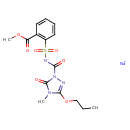
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | Methyl 2-[[[(4,5-dihydro-4-methyl-5-oxo-3-propoxy-1H-1,2,4-triazol-1-yl)carbonyl]amino]sulfonyl]benzoate sodium salt | | Olympus | | Procarbazone sodium | | Procarbazone-sodium | | Sodium N-{[2-(methoxycarbonyl)phenyl]sulfonyl}-4-methyl-5-oxo-3-propoxy-4,5-dihydro-1H-1,2,4-triazole-1-carboximidate | | Sodium {[2-(methoxycarbonyl)phenyl]sulfonyl}[(4-methyl-5-oxo-3-propoxy-4,5-dihydro-1H-1,2,4-triazol-1-yl)carbonyl]azanide |
|
|---|
| Chemical Formula | C15H17N4NaO7S |
|---|
| Average Molecular Mass | 420.373 g/mol |
|---|
| Monoisotopic Mass | 420.072 g/mol |
|---|
| CAS Registry Number | 181274-15-7 |
|---|
| IUPAC Name | sodium [2-(methoxycarbonyl)benzenesulfonyl](4-methyl-5-oxo-3-propoxy-4,5-dihydro-1H-1,2,4-triazole-1-carbonyl)azanide |
|---|
| Traditional Name | sodium [2-(methoxycarbonyl)benzenesulfonyl](4-methyl-5-oxo-3-propoxy-1,2,4-triazole-1-carbonyl)azanide |
|---|
| SMILES | [Na+].CCCOC1=NN(C(=O)[N-]S(=O)(=O)C2=C(C=CC=C2)C(=O)OC)C(=O)N1C |
|---|
| InChI Identifier | InChI=1S/C15H18N4O7S.Na/c1-4-9-26-14-16-19(15(22)18(14)2)13(21)17-27(23,24)11-8-6-5-7-10(11)12(20)25-3;/h5-8H,4,9H2,1-3H3,(H,17,21);/q;+1/p-1 |
|---|
| InChI Key | InChIKey=JRQGDDUXDKCWRF-UHFFFAOYSA-M |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as benzoic acid esters. These are ester derivatives of benzoic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Benzoic acids and derivatives |
|---|
| Direct Parent | Benzoic acid esters |
|---|
| Alternative Parents | |
|---|
| Substituents | - Benzenesulfonamide
- Benzoate ester
- Benzenesulfonyl group
- Benzoyl
- Alkyl aryl ether
- Sulfonylurea
- Azole
- Triazoline
- Heteroaromatic compound
- Organic sulfonic acid or derivatives
- Organosulfonic acid or derivatives
- Sulfonyl
- 1,2,4-triazole
- Methyl ester
- Carboxylic acid ester
- Carbonic acid derivative
- Carboxylic acid derivative
- Azacycle
- Ether
- Monocarboxylic acid or derivatives
- Organic alkali metal salt
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Carbene-type 1,3-dipolar compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organonitrogen compound
- Organooxygen compound
- Organosulfur compound
- Organic sodium salt
- Hydrocarbon derivative
- Organic salt
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Exogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | Not Available |
|---|
| Pathways | Not Available |
|---|
| Applications | Not Available |
|---|
| Biological Roles | Not Available |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | Not Available | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0830-9867300000-a4c38d0e874db4b021ef | 2019-02-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-02tc-5920000000-a5bf54dd30303c368c3e | 2019-02-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052o-9500000000-51b62af008b46ae30deb | 2019-02-22 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4j-9000000000-aa0c8534ae2e09b7f605 | 2019-02-23 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-02ar-8498000000-553cd65ec5ca309474ed | 2019-02-23 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4j-9110000000-4458a7d68921e9ec552e | 2019-02-23 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Not Available |
|---|
| Mechanism of Toxicity | Not Available |
|---|
| Metabolism | Not Available |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | This is a man-made compound that is used as a pesticide. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Not Available |
|---|
| Symptoms | Not Available |
|---|
| Treatment | Not Available |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| HMDB ID | Not Available |
|---|
| PubChem Compound ID | 12056759 |
|---|
| ChEMBL ID | Not Available |
|---|
| ChemSpider ID | 7852353 |
|---|
| KEGG ID | Not Available |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | Not Available |
|---|
| BioCyc ID | Not Available |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | Not Available |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D3911.pdf |
|---|
| General References | Not Available |
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|