| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2014-08-29 05:46:47 UTC |
|---|
| Update Date | 2014-12-24 20:26:40 UTC |
|---|
| Accession Number | T3D4149 |
|---|
| Identification |
|---|
| Common Name | Guanidoacetic acid |
|---|
| Class | Small Molecule |
|---|
| Description | Guanidinoacetic acid is a uremic toxin. Uremic toxins can be subdivided into three major groups based upon their chemical and physical characteristics: 1) small, water-soluble, non-protein-bound compounds, such as urea; 2) small, lipid-soluble and/or protein-bound compounds, such as the phenols and 3) larger so-called middle-molecules, such as beta2-microglobulin. Chronic exposure of uremic toxins can lead to a number of conditions including renal damage, chronic kidney disease and cardiovascular disease.
Guanidoacetic acid is a metabolite in the Urea cycle and metabolism of amino groups, and in the metabolic pathways of several amino acids. This includes glycine, serine, threonine, arginine and proline metabolism. Guanidinoacetic acid is also a precursor of creatine, an essential substrate for muscle energy metabolism. |
|---|
| Compound Type | - Affinity Label
- Amide
- Amine
- Food Toxin
- Lachrymator
- Metabolite
- Natural Compound
- Organic Compound
- Uremic Toxin
|
|---|
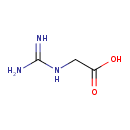
| Chemical Structure | |
|---|
| Synonyms | | Synonym | | (carboxymethyl)-Guanidine | | 2-[[Amino(imino)methyl]amino]acetate | | 2-[[Amino(imino)methyl]amino]acetic acid | | a-Guanidinoacetate | | a-Guanidinoacetic acid | | alpha-Guanidinoacetate | | alpha-Guanidinoacetic acid | | b-Guanidinoacetate | | b-Guanidinoacetic acid | | beta-Guanidinoacetate | | beta-Guanidinoacetic acid | | Betacyamine | | Betasyamine | | Glycocyamine | | Guanidineacetate | | Guanidineacetic acid | | Guanidinoacetate | | Guanidinoacetic acid | | Guanidoacetate | | Guanidylacetate | | Guanidylacetic acid | | Guanyl glycine | | N-Amidino-Glycine | | N-Amidinoglycine | | [(aminoiminomethyl)amino]-Acetate | | [(aminoiminomethyl)amino]-Acetic acid |
|
|---|
| Chemical Formula | C3H7N3O2 |
|---|
| Average Molecular Mass | 117.107 g/mol |
|---|
| Monoisotopic Mass | 117.054 g/mol |
|---|
| CAS Registry Number | 352-97-6 |
|---|
| IUPAC Name | 2-carbamimidamidoacetic acid |
|---|
| Traditional Name | glycocyamine |
|---|
| SMILES | NC(=N)NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H7N3O2/c4-3(5)6-1-2(7)8/h1H2,(H,7,8)(H4,4,5,6) |
|---|
| InChI Key | InChIKey=BPMFZUMJYQTVII-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acids and derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon), or a derivative thereof. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid or derivatives
- Guanidine
- Carboximidamide
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Imine
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Biological Properties |
|---|
| Status | Detected and Not Quantified |
|---|
| Origin | Endogenous |
|---|
| Cellular Locations | |
|---|
| Biofluid Locations | Not Available |
|---|
| Tissue Locations | |
|---|
| Pathways | | Name | SMPDB Link | KEGG Link |
|---|
| Arginine and Proline Metabolism | SMP00020 | map00330 | | Glycine and Serine Metabolism | SMP00004 | map00260 | | Argininemia | SMP00357 | Not Available | | Creatine deficiency, guanidinoacetate methyltransferase deficiency | SMP00504 | Not Available | | Guanidinoacetate Methyltransferase Deficiency (GAMT Deficiency) | SMP00188 | Not Available | | S-Adenosylhomocysteine (SAH) Hydrolase Deficiency | SMP00214 | Not Available |
|
|---|
| Applications | Not Available |
|---|
| Biological Roles | |
|---|
| Chemical Roles | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Appearance | White powder. |
|---|
| Experimental Properties | | Property | Value |
|---|
| Melting Point | Not Available | | Boiling Point | Not Available | | Solubility | 3.6 mg/mL at 15°C | | LogP | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | Deposition Date | View |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-0901000000-4a89fe463d29379e6339 | 2017-09-12 | View Spectrum | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-0911000000-542baca9dcca73225bf0 | 2017-09-12 | View Spectrum | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-006t-0911200000-bb7ea068f193f39c283b | 2017-09-12 | View Spectrum | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-0901000000-4a89fe463d29379e6339 | 2018-05-18 | View Spectrum | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-00di-0911000000-542baca9dcca73225bf0 | 2018-05-18 | View Spectrum | | GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-006t-0911200000-bb7ea068f193f39c283b | 2018-05-18 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9100000000-c703c631a81e400681bd | 2017-08-28 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00dl-9700000000-1dae820000c4f9ec1662 | 2017-10-06 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | 2021-11-05 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-016r-7900000000-4777d20e1ab29262427e | 2012-07-24 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00dl-9000000000-a1b5501bf0f7a9b0489d | 2012-07-24 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0006-9100000000-b61a48cf749b57a2c77e | 2012-07-24 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0gk9-7900000000-5ba298286886c1fabdf5 | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-00di-9100000000-a5b77a913821d637210f | 2012-08-31 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-00di-9100000000-a5b77a913821d637210f | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0gk9-7900000000-5ba298286886c1fabdf5 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-014i-6900000000-4d655a894c702bdfdb59 | 2017-09-14 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-23d5c2795ed49f043127 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-ae360cd8919c8fbce3ee | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0900000000-93406e2cdb79770abc7d | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-00di-9000000000-c6bd00733ea6fa3f6e5d | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-9000000000-2667e0007de6a723415e | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0100-9400000000-c5561d5ccaf260c048ec | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0g29-9400000000-421d076c4b6868ca69fa | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-014i-0900000000-53db4bf2729a2990a938 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-05fu-9000000000-2cad7322e574eb992175 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-01di-9800000000-967331a78fe4a7917408 | 2021-09-20 | View Spectrum | | LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-9000000000-7b281fae734baaa2695d | 2021-09-20 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-4900000000-1b831140a3071dd98e0c | 2017-07-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9300000000-b27aea734be08f5d2ddc | 2017-07-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dl-9000000000-dea2f28faa5430d7e56f | 2017-07-25 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00xr-9500000000-1ba345c7064e9cb70f9c | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9000000000-6864d4355d3462fbc69f | 2017-07-26 | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-1c36299ab8746bbf67a5 | 2017-07-26 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | 2012-12-04 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | Not Available | 2012-12-04 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | 2021-09-29 | View Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 400 MHz, H2O, experimental) | Not Available | 2012-12-04 | View Spectrum |
|
|---|
| Toxicity Profile |
|---|
| Route of Exposure | Endogenous, Ingestion, Dermal (contact) |
|---|
| Mechanism of Toxicity | Uremic toxins such as guanidinoacetic acid are actively transported into the kidneys via organic ion transporters (especially OAT3). Increased levels of uremic toxins can stimulate the production of reactive oxygen species. This seems to be mediated by the direct binding or inhibition by uremic toxins of the enzyme NADPH oxidase (especially NOX4 which is abundant in the kidneys and heart) (2). Reactive oxygen species can induce several different DNA methyltransferases (DNMTs) which are involved in the silencing of a protein known as KLOTHO. KLOTHO has been identified as having important roles in anti-aging, mineral metabolism, and vitamin D metabolism. A number of studies have indicated that KLOTHO mRNA and protein levels are reduced during acute or chronic kidney diseases in response to high local levels of reactive oxygen species (3) |
|---|
| Metabolism | Guanidino acetic acid (GAA) is synthesized in the liver and kidney from Arg and Gly and subsequently methylated by S-adenosylmethionine to form creatine. The conversion of guaniodacetate to creatinine in the liver causes a depletion of methyl groups. This causes homocysteine levels to rise, which has been shown to produce cardiovascular and skeletal problems. |
|---|
| Toxicity Values | Not Available |
|---|
| Lethal Dose | Not Available |
|---|
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). |
|---|
| Uses/Sources | Naturally produced by the body (endogenous). Is used as a supplement to enhance athletic performance. |
|---|
| Minimum Risk Level | Not Available |
|---|
| Health Effects | Acute exposure to guanidoacetic acid can lead to some mild skin and eye irritation. Chronic exposure to uremic toxins can lead to a number of conditions including renal damage, chronic kidney disease and cardiovascular disease. Chronically high levels of guanidoacetic acid are associated with at least 4 inborn errors of metabolism including: Argininemia, Guanidinoacetate methyltransferase deficiency, Guanidinoacetate Methyltransferase Deficiency (GAMT Deficiency) and S-Adenosylhomocysteine (SAH) Hydrolase Deficiency. |
|---|
| Symptoms | As a uremic toxin, this compound can cause uremic syndrome. Uremic syndrome may affect any part of the body and can cause nausea, vomiting, loss of appetite, and weight loss. It can also cause changes in mental status, such as confusion, reduced awareness, agitation, psychosis, seizures, and coma. Abnormal bleeding, such as bleeding spontaneously or profusely from a very minor injury can also occur. Heart problems, such as an irregular heartbeat, inflammation in the sac that surrounds the heart (pericarditis), and increased pressure on the heart can be seen in patients with uremic syndrome. Shortness of breath from fluid buildup in the space between the lungs and the chest wall (pleural effusion) can also be present. |
|---|
| Treatment | Chronic Exposure: Kidney dialysis is usually needed to relieve the symptoms of uremic syndrome until normal kidney function can be restored.
Acute Exposure: EYES: irrigate opened eyes for several minutes under running water. INGESTION: do not induce vomiting. Rinse mouth with water (never give anything by mouth to an unconscious person). Seek immediate medical advice. |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| External Links |
|---|
| DrugBank ID | DB02751 |
|---|
| HMDB ID | HMDB00128 |
|---|
| PubChem Compound ID | 763 |
|---|
| ChEMBL ID | CHEMBL281593 |
|---|
| ChemSpider ID | 743 |
|---|
| KEGG ID | C00581 |
|---|
| UniProt ID | Not Available |
|---|
| OMIM ID | |
|---|
| ChEBI ID | 16344 |
|---|
| BioCyc ID | GUANIDOACETIC_ACID |
|---|
| CTD ID | Not Available |
|---|
| Stitch ID | Not Available |
|---|
| PDB ID | NMG |
|---|
| ACToR ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| MSDS | T3D4149.pdf |
|---|
| General References | - Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A: Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012 Jul;23(7):1258-70. doi: 10.1681/ASN.2011121175. Epub 2012 May 24. [22626821 ]
- Schulz AM, Terne C, Jankowski V, Cohen G, Schaefer M, Boehringer F, Tepel M, Kunkel D, Zidek W, Jankowski J: Modulation of NADPH oxidase activity by known uraemic retention solutes. Eur J Clin Invest. 2014 Aug;44(8):802-11. doi: 10.1111/eci.12297. [25041433 ]
- Young GH, Wu VC: KLOTHO methylation is linked to uremic toxins and chronic kidney disease. Kidney Int. 2012 Apr;81(7):611-2. doi: 10.1038/ki.2011.461. [22419041 ]
- Schulze A: Creatine deficiency syndromes. Mol Cell Biochem. 2003 Feb;244(1-2):143-50. [12701824 ]
- Mercimek-Mahmutoglu S, Stoeckler-Ipsiroglu S, Adami A, Appleton R, Araujo HC, Duran M, Ensenauer R, Fernandez-Alvarez E, Garcia P, Grolik C, Item CB, Leuzzi V, Marquardt I, Muhl A, Saelke-Kellermann RA, Salomons GS, Schulze A, Surtees R, van der Knaap MS, Vasconcelos R, Verhoeven NM, Vilarinho L, Wilichowski E, Jakobs C: GAMT deficiency: features, treatment, and outcome in an inborn error of creatine synthesis. Neurology. 2006 Aug 8;67(3):480-4. Epub 2006 Jul 19. [16855203 ]
- Isbrandt D, von Figura K: Cloning and sequence analysis of human guanidinoacetate N-methyltransferase cDNA. Biochim Biophys Acta. 1995 Dec 27;1264(3):265-7. [8547310 ]
- Ensenauer R, Thiel T, Schwab KO, Tacke U, Stockler-Ipsiroglu S, Schulze A, Hennig J, Lehnert W: Guanidinoacetate methyltransferase deficiency: differences of creatine uptake in human brain and muscle. Mol Genet Metab. 2004 Jul;82(3):208-13. [15234333 ]
- Yasuda M, Sugahara K, Zhang J, Ageta T, Nakayama K, Shuin T, Kodama H: Simultaneous determination of creatinine, creatine, and guanidinoacetic acid in human serum and urine using liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. Anal Biochem. 1997 Nov 15;253(2):231-5. [9367508 ]
- Sijens PE, Reijngoud DJ, Soorani-Lunsing RJ, Oudkerk M, van Spronsen FJ: Cerebral 1H MR spectroscopy showing elevation of brain guanidinoacetate in argininosuccinate lyase deficiency. Mol Genet Metab. 2006 May;88(1):100-2. Epub 2005 Dec 15. [16343968 ]
- Valongo C, Cardoso ML, Domingues P, Almeida L, Verhoeven N, Salomons G, Jakobs C, Vilarinho L: Age related reference values for urine creatine and guanidinoacetic acid concentration in children and adolescents by gas chromatography-mass spectrometry. Clin Chim Acta. 2004 Oct;348(1-2):155-61. [15369749 ]
- Fingerhut R: Stable isotope dilution method for the determination of guanidinoacetic acid by gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(7):717-22. [12661026 ]
- Schmidt A, Marescau B, Boehm EA, Renema WK, Peco R, Das A, Steinfeld R, Chan S, Wallis J, Davidoff M, Ullrich K, Waldschutz R, Heerschap A, De Deyn PP, Neubauer S, Isbrandt D: Severely altered guanidino compound levels, disturbed body weight homeostasis and impaired fertility in a mouse model of guanidinoacetate N-methyltransferase (GAMT) deficiency. Hum Mol Genet. 2004 May 1;13(9):905-21. Epub 2004 Mar 17. [15028668 ]
- Stockler S, Isbrandt D, Hanefeld F, Schmidt B, von Figura K: Guanidinoacetate methyltransferase deficiency: the first inborn error of creatine metabolism in man. Am J Hum Genet. 1996 May;58(5):914-22. [8651275 ]
- Shirokane Y, Utsushikawa M, Nakajima M: A new enzymic determination of guanidinoacetic acid in urine. Clin Chem. 1987 Mar;33(3):394-7. [3815805 ]
- Mizutani N, Hayakawa C, Ohya Y, Watanabe K, Watanabe Y, Mori A: Guanidino compounds in hyperargininemia. Tohoku J Exp Med. 1987 Nov;153(3):197-205. [3433275 ]
- Schulze A, Ebinger F, Rating D, Mayatepek E: Improving treatment of guanidinoacetate methyltransferase deficiency: reduction of guanidinoacetic acid in body fluids by arginine restriction and ornithine supplementation. Mol Genet Metab. 2001 Dec;74(4):413-9. [11749046 ]
- Stockler S, Holzbach U, Hanefeld F, Marquardt I, Helms G, Requart M, Hanicke W, Frahm J: Creatine deficiency in the brain: a new, treatable inborn error of metabolism. Pediatr Res. 1994 Sep;36(3):409-13. [7808840 ]
|
|---|
| Gene Regulation |
|---|
| Up-Regulated Genes | Not Available |
|---|
| Down-Regulated Genes | Not Available |
|---|