Propazine (T3D3908)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2013-04-25 07:56:54 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:34 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D3908 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Propazine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Propazine is an herbicide used for control of broadleaf weeds and annual grasses in sweet sorghum. It is applied as a spray at the time of planting or immediately following planting, but prior to weed or sorghum emergence. It is also used as a postemergence selective herbicide on carrots, celery and fennel. Propazine is classified as a slightly toxic compound in EPA toxicity class III and as a General Use Pesticide. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
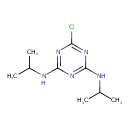
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C9H16ClN5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 229.710 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 229.109 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 139-40-2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 6-chloro-N2,N4-bis(propan-2-yl)-1,3,5-triazine-2,4-diamine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | propazine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CC(C)NC1=NC(NC(C)C)=NC(Cl)=N1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C9H16ClN5/c1-5(2)11-8-13-7(10)14-9(15-8)12-6(3)4/h5-6H,1-4H3,(H2,11,12,13,14,15) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=WJNRPILHGGKWCK-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 1,3,5-triazine-2,4-diamines. These are aromatic compounds containing a 1,3,5-triazine ring which is 2,4-disusbtituted wit amine groups. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Triazines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Aminotriazines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 1,3,5-triazine-2,4-diamines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | This is a man-made compound that is used as a pesticide. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 4937 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL1333189 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 4768 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C14312 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 38067 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D3908.pdf | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Most active in catalyzing 2-hydroxylation. Caffeine is metabolized primarily by cytochrome CYP1A2 in the liver through an initial N3-demethylation. Also acts in the metabolism of aflatoxin B1 and acetaminophen. Participates in the bioactivation of carcinogenic aromatic and heterocyclic amines. Catalizes the N-hydroxylation of heterocyclic amines and the O-deethylation of phenacetin.
- Gene Name:
- CYP1A2
- Uniprot ID:
- P05177
- Molecular Weight:
- 58293.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.58 uM | CLZD_CYP1A2_24 | CellzDirect |
| AC50 | 5.97 uM | CLZD_CYP1A2_48 | CellzDirect |
| AC50 | 0.64 uM | CLZD_CYP1A2_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Metal ion binding
- Specific Function:
- Hydrolyzes the second messenger cAMP, which is a key regulator of many important physiological processes.
- Gene Name:
- PDE4A
- Uniprot ID:
- P27815
- Molecular Weight:
- 98142.155 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.05 uM | NVS_ENZ_hPDE4A1 | Novascreen |
| AC50 | 5.10 uM | NVS_ENZ_hPDE4A1 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- CCL2
- Uniprot ID:
- P13500
- Molecular Weight:
- 11024.87 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_SAg_MCP1_down | BioSeek |
| AC50 | 1.48 uM | BSK_3C_MCP1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Virus receptor activity
- Specific Function:
- ICAM proteins are ligands for the leukocyte adhesion protein LFA-1 (integrin alpha-L/beta-2). During leukocyte trans-endothelial migration, ICAM1 engagement promotes the assembly of endothelial apical cups through ARHGEF26/SGEF and RHOG activation.(Microbial infection) Acts as a receptor for major receptor group rhinovirus A-B capsid proteins (PubMed:1968231, PubMed:2538243). Acts as a receptor for Coxsackievirus A21 capsid proteins (PubMed:11160747, PubMed:16004874, PubMed:9539703). Upon Kaposi's sarcoma-associated herpesvirus/HHV-8 infection, is degraded by viral E3 ubiquitin ligase MIR2, presumably to prevent lysis of infected cells by cytotoxic T-lymphocytes and NK cell (PubMed:11413168).
- Gene Name:
- ICAM1
- Uniprot ID:
- P05362
- Molecular Weight:
- 57824.785 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_KF3CT_ICAM1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.03 uM | CLZD_CYP2B6_6 | CellzDirect |
| AC50 | 6.84 uM | CLZD_CYP2B6_24 | CellzDirect |
| AC50 | 4.06 uM | CLZD_CYP2B6_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Cell-surface glycoprotein having a role in immunoadhesion. Mediates in the adhesion of blood neutrophils in cytokine-activated endothelium through interaction with PSGL1/SELPLG. May have a role in capillary morphogenesis.
- Gene Name:
- SELE
- Uniprot ID:
- P16581
- Molecular Weight:
- 66654.575 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_Eselectin_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Cytokine activity
- Specific Function:
- Produced by activated macrophages, IL-1 stimulates thymocyte proliferation by inducing IL-2 release, B-cell maturation and proliferation, and fibroblast growth factor activity. IL-1 proteins are involved in the inflammatory response, being identified as endogenous pyrogens, and are reported to stimulate the release of prostaglandin and collagenase from synovial cells.
- Gene Name:
- IL1A
- Uniprot ID:
- P01583
- Molecular Weight:
- 30606.29 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_BE3C_IL1a_up | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Tumor necrosis factor receptor binding
- Specific Function:
- Cytokine that binds to TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. It is mainly secreted by macrophages and can induce cell death of certain tumor cell lines. It is potent pyrogen causing fever by direct action or by stimulation of interleukin-1 secretion and is implicated in the induction of cachexia, Under certain conditions it can stimulate cell proliferation and induce cell differentiation. Impairs regulatory T-cells (Treg) function in individuals with rheumatoid arthritis via FOXP3 dephosphorylation. Upregulates the expression of protein phosphatase 1 (PP1), which dephosphorylates the key 'Ser-418' residue of FOXP3, thereby inactivating FOXP3 and rendering Treg cells functionally defective (PubMed:23396208). Key mediator of cell death in the anticancer action of BCG-stimulated neutrophils in combination with DIABLO/SMAC mimetic in the RT4v6 bladder cancer cell line (PubMed:22517918).The TNF intracellular domain (ICD) form induces IL12 production in dendritic cells.
- Gene Name:
- TNF
- Uniprot ID:
- P01375
- Molecular Weight:
- 25644.15 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_LPS_TNFa_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 7.80 uM | CLZD_CYP3A4_6 | CellzDirect |
| AC50 | 5.66 uM | CLZD_CYP3A4_24 | CellzDirect |
| AC50 | 7.11 uM | CLZD_CYP3A4_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid binding
- Specific Function:
- UDPGT is of major importance in the conjugation and subsequent elimination of potentially toxic xenobiotics and endogenous compounds. This isoform glucuronidates bilirubin IX-alpha to form both the IX-alpha-C8 and IX-alpha-C12 monoconjugates and diconjugate. Is also able to catalyze the glucuronidation of 17beta-estradiol, 17alpha-ethinylestradiol, 1-hydroxypyrene, 4-methylumbelliferone, 1-naphthol, paranitrophenol, scopoletin, and umbelliferone. Isoform 2 lacks transferase activity but acts as a negative regulator of isoform 1.
- Gene Name:
- UGT1A1
- Uniprot ID:
- P22309
- Molecular Weight:
- 59590.91 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.72 uM | CLZD_UGT1A1_24 | CellzDirect |
| AC50 | 6.71 uM | CLZD_UGT1A1_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Vitamin d 24-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP1A1
- Uniprot ID:
- P04798
- Molecular Weight:
- 58164.815 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 5.76 uM | CLZD_CYP1A1_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]