Butafenacil (T3D3803)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2013-04-25 07:56:50 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:26:32 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D3803 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Butafenacil | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Butafenacil is a herbicide belonging to the pyrimidindione chemical class. Butafenacil is intended for use as a cotton defoliant. Subchronic toxicity caused by butafenacil is characterized primarily by hematological effects such as, decreased hemoglobin, hematocrit, mean corpuscular hemoglobin (MCH), and mean corpuscular volume (MCV), and increased red cell volume. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
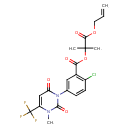
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C20H18ClF3N2O6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 474.815 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 474.081 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 134605-64-4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | prop-2-en-1-yl 2-{2-chloro-5-[3-methyl-2,6-dioxo-4-(trifluoromethyl)-1,2,3,6-tetrahydropyrimidin-1-yl]benzoyloxy}-2-methylpropanoate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | prop-2-en-1-yl 2-{2-chloro-5-[3-methyl-2,6-dioxo-4-(trifluoromethyl)pyrimidin-1-yl]benzoyloxy}-2-methylpropanoate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CN1C(=O)N(C(=O)C=C1C(F)(F)F)C1=CC(C(=O)OC(C)(C)C(=O)OCC=C)=C(Cl)C=C1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C20H18ClF3N2O6/c1-5-8-31-17(29)19(2,3)32-16(28)12-9-11(6-7-13(12)21)26-15(27)10-14(20(22,23)24)25(4)18(26)30/h5-7,9-10H,1,8H2,2-4H3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=JEDYYFXHPAIBGR-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as benzoic acid esters. These are ester derivatives of benzoic acid. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Benzenoids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Benzene and substituted derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Benzoic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Benzoic acid esters | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | White powder. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | This is a man-made compound that is used as a pesticide. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 11826859 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL1867342 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 10001509 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D3803.pdf | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Vitamin d3 25-hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It performs a variety of oxidation reactions (e.g. caffeine 8-oxidation, omeprazole sulphoxidation, midazolam 1'-hydroxylation and midazolam 4-hydroxylation) of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,8-cineole 2-exo-monooxygenase. The enzyme also hydroxylates etoposide (PubMed:11159812). Catalyzes 4-beta-hydroxylation of cholesterol. May catalyze 25-hydroxylation of cholesterol in vitro (PubMed:21576599).
- Gene Name:
- CYP3A4
- Uniprot ID:
- P08684
- Molecular Weight:
- 57342.67 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.30 uM | CLZD_CYP3A4_6 | CellzDirect |
| AC50 | 0.51 uM | CLZD_CYP3A4_24 | CellzDirect |
| AC50 | 3.87 uM | CLZD_CYP3A4_48 | CellzDirect |
| AC50 | 0.30 uM | CLZD_CYP3A4_6 | CellzDirect |
| AC50 | 0.51 uM | CLZD_CYP3A4_24 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Leukotriene-b4 20-monooxygenase activity
- Specific Function:
- Catalyzes leukotriene B4 omega-hydroxylation and arachidonic acid omega-hydroxylation but with an activity much lower than that of CYP4F2. Catalyzes the hydroxylation of the antihistamine ebastine.
- Gene Name:
- CYP4F12
- Uniprot ID:
- Q9HCS2
- Molecular Weight:
- 60269.165 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.38 uM | NVS_ADME_hCYP4F12_Activator | Novascreen |
| AC50 | 0.37 uM | NVS_ADME_hCYP4F12_Activator | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 0.72 uM | CLZD_CYP2B6_6 | CellzDirect |
| AC50 | 0.43 uM | CLZD_CYP2B6_24 | CellzDirect |
| AC50 | 0.40 uM | CLZD_CYP2B6_48 | CellzDirect |
| AC50 | 0.72 uM | CLZD_CYP2B6_6 | CellzDirect |
| AC50 | 0.43 uM | CLZD_CYP2B6_24 | CellzDirect |
| AC50 | 0.40 uM | CLZD_CYP2B6_48 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Oxygen binding
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP3A5
- Uniprot ID:
- P20815
- Molecular Weight:
- 57108.065 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.80 uM | NVS_ADME_hCYP3A5 | Novascreen |
| AC50 | 0.60 uM | NVS_ADME_hCYP3A5 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear receptor that binds and is activated by variety of endogenous and xenobiotic compounds. Transcription factor that activates the transcription of multiple genes involved in the metabolism and secretion of potentially harmful xenobiotics, drugs and endogenous compounds. Activated by the antibiotic rifampicin and various plant metabolites, such as hyperforin, guggulipid, colupulone, and isoflavones. Response to specific ligands is species-specific. Activated by naturally occurring steroids, such as pregnenolone and progesterone. Binds to a response element in the promoters of the CYP3A4 and ABCB1/MDR1 genes.
- Gene Name:
- NR1I2
- Uniprot ID:
- O75469
- Molecular Weight:
- 49761.245 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 2.40 uM | NVS_NR_hPXR | Novascreen |
| AC50 | 9.86 uM | ATG_PXRE_CIS | Attagene |
| AC50 | 0.73 uM | NVS_NR_hPXR | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii core promoter proximal region sequence-specific binding
- Specific Function:
- Endoplasmic reticulum (ER)-bound transcription factor that plays a role in the unfolded protein response (UPR). Involved in cell proliferation and migration, tumor suppression and inflammatory gene expression. Plays also a role in the human immunodeficiency virus type 1 (HIV-1) virus protein expression and in the herpes simplex virus-1 (HSV-1) latent infection and reactivation from latency. Isoform 2 plays a role in the unfolded protein response (UPR). Isoform 2 acts as a positive regulator of LKN-1/CCL15-induced chemotaxis signaling of leukocyte cell migration. Isoform 2 may play a role as a cellular tumor suppressor that is targeted by the hepatitis C virus (HSV) core protein. Isoform 2 represses the VP16-mediated transactivation of immediate early genes of the HSV-1 virus by sequestring host cell factor-1 HCFC1 in the ER membrane of sensory neurons, thereby preventing the initiation of the replicative cascade leading to latent infection. Isoform 3 functions as a negative transcriptional regulator in ligand-induced transcriptional activation of the glucocorticoid receptor NR3C1 by recruiting and activating histone deacetylases (HDAC1, HDAC2 and HDAC6). Isoform 3 decreases the acetylation level of histone H4. Isoform 3 does not promote the chemotactic activity of leukocyte cells.Processed cyclic AMP-responsive element-binding protein 3: acts as a transcription factor that activates unfolded protein response (UPR) target genes during endoplasmic reticulum (ER) stress response. Promotes cell survival against ER stress-induced apoptotic cell death during UPR. Activates transcription from CRE and C/EBP-containing reporter genes. Induces transcriptional activation of chemokine receptors. Activates transcription of genes required for reactivation of the latent HSV-1 virus. Down-regulates Tat-dependent transcription of the HIV-1 LTR by interacting with HIV-1 Tat. It's transcriptional activity is inhibited by CREBZF in a HCFC1-dependent manner, by the viral transactivator protein VP16 and by the HCV core protein. Binds DNA to the cAMP response element (CRE) (consensus: 5'-GTGACGT[AG][AG]-3') and C/EBP sequences present in many viral and cellular promoters. Binds to the unfolded protein respons element (UPRE) consensus sequences sites. Binds DNA to the 5'-CCAC[GA]-3'half of ERSE II (5'-ATTGG-N-CCACG-3'). Associates with chromatin to the HERPUD1 promoter.
- Gene Name:
- CREB3
- Uniprot ID:
- O43889
- Molecular Weight:
- 43916.65 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.31 uM | ATG_CRE_CIS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Not Available
- Specific Function:
- Not Available
- Gene Name:
- CCL2
- Uniprot ID:
- P13500
- Molecular Weight:
- 11024.87 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_KF3CT_MCP1_down | BioSeek |
| AC50 | 1.48 uM | BSK_SM3C_MCP1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Receptor binding
- Specific Function:
- Chemotactic for monocytes and T-lymphocytes. Binds to CXCR3.
- Gene Name:
- CXCL10
- Uniprot ID:
- P02778
- Molecular Weight:
- 10880.915 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_IP10_down | BioSeek |
| AC50 | 1.48 uM | BSK_KF3CT_IP10_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Cleaves collagens of types I, II, and III at one site in the helical domain. Also cleaves collagens of types VII and X. In case of HIV infection, interacts and cleaves the secreted viral Tat protein, leading to a decrease in neuronal Tat's mediated neurotoxicity.
- Gene Name:
- MMP1
- Uniprot ID:
- P03956
- Molecular Weight:
- 54006.61 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_MMP1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Protein homodimerization activity
- Specific Function:
- Cytokine that plays an essential role in the regulation of survival, proliferation and differentiation of hematopoietic precursor cells, especially mononuclear phagocytes, such as macrophages and monocytes. Promotes the release of proinflammatory chemokines, and thereby plays an important role in innate immunity and in inflammatory processes. Plays an important role in the regulation of osteoclast proliferation and differentiation, the regulation of bone resorption, and is required for normal bone development. Required for normal male and female fertility. Promotes reorganization of the actin cytoskeleton, regulates formation of membrane ruffles, cell adhesion and cell migration. Plays a role in lipoprotein clearance.
- Gene Name:
- CSF1
- Uniprot ID:
- P09603
- Molecular Weight:
- 60178.885 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_MCSF_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Serine-type endopeptidase inhibitor activity
- Specific Function:
- Serine protease inhibitor. This inhibitor acts as 'bait' for tissue plasminogen activator, urokinase, protein C and matriptase-3/TMPRSS7. Its rapid interaction with PLAT may function as a major control point in the regulation of fibrinolysis.
- Gene Name:
- SERPINE1
- Uniprot ID:
- P05121
- Molecular Weight:
- 45059.695 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_hDFCGF_PAI1_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Urokinase plasminogen activator receptor activity
- Specific Function:
- Acts as a receptor for urokinase plasminogen activator. Plays a role in localizing and promoting plasmin formation. Mediates the proteolysis-independent signal transduction activation effects of U-PA. It is subject to negative-feedback regulation by U-PA which cleaves it into an inactive form.
- Gene Name:
- PLAUR
- Uniprot ID:
- Q03405
- Molecular Weight:
- 36977.62 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 1.48 uM | BSK_3C_uPAR_down | BioSeek |
| AC50 | 1.48 uM | BSK_SM3C_uPAR_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transferase activity
- Specific Function:
- Synthesizes the second messagers cyclic ADP-ribose and nicotinate-adenine dinucleotide phosphate, the former a second messenger for glucose-induced insulin secretion. Also has cADPr hydrolase activity. Also moonlights as a receptor in cells of the immune system.
- Gene Name:
- CD38
- Uniprot ID:
- P28907
- Molecular Weight:
- 34328.145 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_CD38_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Cytokine activity
- Specific Function:
- Cytokine that affects the growth, movement, or activation state of cells that participate in immune and inflammatory response. Chemotactic for activated T-cells. Binds to CXCR3.
- Gene Name:
- CXCL9
- Uniprot ID:
- Q07325
- Molecular Weight:
- 14018.72 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_hDFCGF_MIG_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Involved in lymphocyte proliferation and functions as a signal transmitting receptor in lymphocytes, natural killer (NK) cells, and platelets.
- Gene Name:
- CD69
- Uniprot ID:
- Q07108
- Molecular Weight:
- 22559.25 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_CD69_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Ubiquitin protein ligase binding
- Specific Function:
- Receptor for TNFSF5/CD40LG. Transduces TRAF6- and MAP3K8-mediated signals that activate ERK in macrophages and B cells, leading to induction of immunoglobulin secretion.
- Gene Name:
- CD40
- Uniprot ID:
- P25942
- Molecular Weight:
- 30618.76 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_CD40_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]