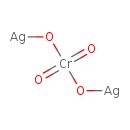
Silver chromate (T3D0709)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-03-26 21:10:18 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:22:37 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D0709 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Silver chromate | ||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | ||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Silver chromate is a chemical compound of silver and hexavalent chromium. It is used in photography. Hexavalent chromium refers to chemical compounds that contain the element chromium in the +6 oxidation state. Chromium(VI) is more toxic than other oxidation states of the chromium atom because of its greater ability to enter cells and higher redox potential. Silver is a metallic element with the chemical symbol Ag and atomic number 47. It occurs naturally in its pure, free form, as an alloy with gold and other metals, and in minerals such as argentite and chlorargyrite. (14, 15, 10, 11) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | |||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | Ag2CrO4 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 331.730 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 329.730 g/mol | ||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 7784-01-2 | ||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | dioxo-2,4-dioxa-1,5-diargenta-3-chromapentane | ||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | dioxo-2,4-dioxa-1,5-diargenta-3-chromapentane | ||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [Ag]O[Cr](=O)(=O)O[Ag] | ||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/2Ag.Cr.4O | ||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=GWVVXXOAOVGAEF-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of inorganic compounds known as transition metal chromates. These are inorganic compounds in which the largest oxoanion is chromate, and in which the heaviest atom is a transition metal. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Inorganic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Mixed metal/non-metal compounds | ||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Transition metal oxoanionic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Transition metal chromates | ||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Transition metal chromates | ||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | ||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Red-brown powder. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | |||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Inhalation (10) ; oral (10) ; dermal (10) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Hexavalent chromium's carcinogenic effects are caused by its metabolites, pentavalent and trivalent chromium. The DNA damage may be caused by hydroxyl radicals produced during reoxidation of pentavalent chromium by hydrogen peroxide molecules present in the cell. Trivalent chromium may also form complexes with peptides, proteins, and DNA, resulting in DNA-protein crosslinks, DNA strand breaks, DNA-DNA interstrand crosslinks, chromium-DNA adducts, chromosomal aberrations and alterations in cellular signaling pathways. It has been shown to induce carcinogenesis by overstimulating cellular regulatory pathways and increasing peroxide levels by activating certain mitogen-activated protein kinases. It can also cause transcriptional repression by cross-linking histone deacetylase 1-DNA methyltransferase 1 complexes to CYP1A1 promoter chromatin, inhibiting histone modification. Chromium may increase its own toxicity by modifying metal regulatory transcription factor 1, causing the inhibition of zinc-induced metallothionein transcription. Metallic silver is oxidized and may deposit in the tissues, causing arygria. The silver ion is known to inhibit glutathione peroxidase and NA+,K+-ATPase activity, disrupting selenium-catalyzed sulfhydryl oxidation-reduction reactions and intracellular ion concentrations, respectively. Silver nanoparticles are believed to disrupt the mitochondrial respiratory chain, causing oxidative stress, reduced ATP synthesis, and DNA damage. (14, 6, 7, 8, 9, 1, 10, 2, 3, 4) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Chromium is absorbed from oral, inhalation, or dermal exposure and distributes to nearly all tissues, with the highest concentrations found in kidney and liver. Bone is also a major storage site and may contribute to long-term retention. Hexavalent chromium's similarity to sulfate and chromate allow it to be transported into cells via sulfate transport mechanisms. Inside the cell, hexavalent chromium is reduced first to pentavalent chromium, then to trivalent chromium by many substances including ascorbate, glutathione, and nicotinamide adenine dinucleotide. Chromium is almost entirely excreted with the urine. Silver compounds can also be absorbed orally and dermally. It distributes throughout the body in the blood, particularily to the liver. Insoluble silver salts are transformed into soluble silver sulfide albuminates, bind to amino or carboxyl groups in RNA, DNA, and proteins, or are reduced to metallic silver by ascorbic acid or catecholamines. Metallic silver is oxidized and may deposit in the tissues, causing arygria. Silver is eliminated primarily in the faeces. (14, 1, 10) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | 1 to 3 grams for an adult human (hexavalent chromium). (5) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | 1, carcinogenic to humans. (13) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Silver chromate is used in photography. (11) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Intermediate Oral: 0.005 mg/kg/day (12) Chronic Oral: 0.001 mg/kg/day (12) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Hexavalent chromium is a known carcinogen. Chronic inhalation especially has been linked to lung cancer. Hexavalent chromium has also been know to cause reproductive and developmental defects. Exposure to high levels of silver for a long period of time may result in a condition called arygria, a blue-gray discoloration of the skin and other body tissues. Argyria is a permanent effect but does not appear to be harmful to health. While silver itself is not toxic, most silver salts are, and may damage the liver, kidney, and central nervous system, as well as be carcinogenic. (14, 15, 16, 1) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Breathing hexavalent chromium can cause irritation to the lining of the nose, nose ulcers, runny nose, and breathing problems, such as asthma, cough, shortness of breath, or wheezing. Ingestion of hexavalent chromium causes irritation and ulcers in the stomach and small intestine, as well as anemia. Skin contact can cause skin ulcers. Exposure to high levels of silver for a long period of time may result in a condition called arygria, a blue-gray discoloration of the skin and other body tissues. Argyria is a permanent effect but does not appear to be harmful to health. Exposure to high levels of silver in the air has resulted in breathing problems, lung and throat irritation, and stomach pains. Skin contact with silver can cause mild allergic reactions such as rash, swelling, and inflammation in some people. (14, 10) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | There is no know antidote for chromium poisoning. Exposure is usually handled with symptomatic treatment. (10) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 62666 | ||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | |||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | C038224 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Silver chromate | ||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | T3D0709.pdf | ||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | |||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||
Targets
1. DNA
- General Function:
- Used for biological information storage.
- Specific Function:
- DNA contains the instructions needed for an organism to develop, survive and reproduce.
- Molecular Weight:
- 2.15 x 1012 Da
References
- ATSDR - Agency for Toxic Substances and Disease Registry (2008). Toxicological profile for chromium. U.S. Public Health Service in collaboration with U.S. Environmental Protection Agency (EPA). [Link]
- General Function:
- Glutathione peroxidase activity
- Specific Function:
- Protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide, by glutathione. May constitute a glutathione peroxidase-like protective system against peroxide damage in sperm membrane lipids.
- Gene Name:
- GPX5
- Uniprot ID:
- O75715
- Molecular Weight:
- 25202.14 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Sh3 domain binding
- Specific Function:
- Protects the hemoglobin in erythrocytes from oxidative breakdown.
- Gene Name:
- GPX1
- Uniprot ID:
- P07203
- Molecular Weight:
- 22087.94 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Glutathione peroxidase activity
- Specific Function:
- Could play a major role in protecting mammals from the toxicity of ingested organic hydroperoxides. Tert-butyl hydroperoxide, cumene hydroperoxide and linoleic acid hydroperoxide but not phosphatidycholine hydroperoxide, can act as acceptors.
- Gene Name:
- GPX2
- Uniprot ID:
- P18283
- Molecular Weight:
- 21953.835 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Transcription factor binding
- Specific Function:
- Protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide, by glutathione.
- Gene Name:
- GPX3
- Uniprot ID:
- P22352
- Molecular Weight:
- 25552.185 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Glutathione peroxidase activity
- Specific Function:
- Not Available
- Gene Name:
- GPX6
- Uniprot ID:
- P59796
- Molecular Weight:
- 24970.46 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Peroxidase activity
- Specific Function:
- It protects esophageal epithelia from hydrogen peroxide-induced oxidative stress. It suppresses acidic bile acid-induced reactive oxigen species (ROS) and protects against oxidative DNA damage and double-strand breaks.
- Gene Name:
- GPX7
- Uniprot ID:
- Q96SL4
- Molecular Weight:
- 20995.88 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Transcription regulatory region sequence-specific dna binding
- Specific Function:
- Responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone deacetylation gives a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. Histone deacetylases act via the formation of large multiprotein complexes. Deacetylates SP proteins, SP1 and SP3, and regulates their function. Component of the BRG1-RB1-HDAC1 complex, which negatively regulates the CREST-mediated transcription in resting neurons. Upon calcium stimulation, HDAC1 is released from the complex and CREBBP is recruited, which facilitates transcriptional activation. Deacetylates TSHZ3 and regulates its transcriptional repressor activity. Deacetylates 'Lys-310' in RELA and thereby inhibits the transcriptional activity of NF-kappa-B. Deacetylates NR1D2 and abrogates the effect of KAT5-mediated relieving of NR1D2 transcription repression activity. Component of a RCOR/GFI/KDM1A/HDAC complex that suppresses, via histone deacetylase (HDAC) recruitment, a number of genes implicated in multilineage blood cell development. Involved in CIART-mediated transcriptional repression of the circadian transcriptional activator: CLOCK-ARNTL/BMAL1 heterodimer. Required for the transcriptional repression of circadian target genes, such as PER1, mediated by the large PER complex or CRY1 through histone deacetylation.
- Gene Name:
- HDAC1
- Uniprot ID:
- Q13547
- Molecular Weight:
- 55102.615 Da
References
- Schnekenburger M, Talaska G, Puga A: Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol. 2007 Oct;27(20):7089-101. Epub 2007 Aug 6. [17682057 ]
- General Function:
- Transcriptional activator activity, rna polymerase ii core promoter proximal region sequence-specific binding
- Specific Function:
- Activates the metallothionein I promoter. Binds to the metal responsive element (MRE).
- Gene Name:
- MTF1
- Uniprot ID:
- Q14872
- Molecular Weight:
- 80956.22 Da
References
- Kimura T: [Molecular mechanism involved in chromium(VI) toxicity]. Yakugaku Zasshi. 2007 Dec;127(12):1957-65. [18057785 ]
- General Function:
- Rna polymerase ii carboxy-terminal domain kinase activity
- Specific Function:
- Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, DCC, FRS2 or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3 or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6 or DUSP16) are other substrates which enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade. Mediates phosphorylation of TPR in respons to EGF stimulation. May play a role in the spindle assembly checkpoint. Phosphorylates PML and promotes its interaction with PIN1, leading to PML degradation.Acts as a transcriptional repressor. Binds to a [GC]AAA[GC] consensus sequence. Repress the expression of interferon gamma-induced genes. Seems to bind to the promoter of CCL5, DMP1, IFIH1, IFITM1, IRF7, IRF9, LAMP3, OAS1, OAS2, OAS3 and STAT1. Transcriptional activity is independent of kinase activity.
- Gene Name:
- MAPK1
- Uniprot ID:
- P28482
- Molecular Weight:
- 41389.265 Da
References
- Kim G, Yurkow EJ: Chromium induces a persistent activation of mitogen-activated protein kinases by a redox-sensitive mechanism in H4 rat hepatoma cells. Cancer Res. 1996 May 1;56(9):2045-51. [8616849 ]
- General Function:
- Phosphatase binding
- Specific Function:
- Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. MAPK1/ERK2 and MAPK3/ERK1 are the 2 MAPKs which play an important role in the MAPK/ERK cascade. They participate also in a signaling cascade initiated by activated KIT and KITLG/SCF. Depending on the cellular context, the MAPK/ERK cascade mediates diverse biological functions such as cell growth, adhesion, survival and differentiation through the regulation of transcription, translation, cytoskeletal rearrangements. The MAPK/ERK cascade plays also a role in initiation and regulation of meiosis, mitosis, and postmitotic functions in differentiated cells by phosphorylating a number of transcription factors. About 160 substrates have already been discovered for ERKs. Many of these substrates are localized in the nucleus, and seem to participate in the regulation of transcription upon stimulation. However, other substrates are found in the cytosol as well as in other cellular organelles, and those are responsible for processes such as translation, mitosis and apoptosis. Moreover, the MAPK/ERK cascade is also involved in the regulation of the endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC); as well as in the fragmentation of the Golgi apparatus during mitosis. The substrates include transcription factors (such as ATF2, BCL6, ELK1, ERF, FOS, HSF4 or SPZ1), cytoskeletal elements (such as CANX, CTTN, GJA1, MAP2, MAPT, PXN, SORBS3 or STMN1), regulators of apoptosis (such as BAD, BTG2, CASP9, DAPK1, IER3, MCL1 or PPARG), regulators of translation (such as EIF4EBP1) and a variety of other signaling-related molecules (like ARHGEF2, FRS2 or GRB10). Protein kinases (such as RAF1, RPS6KA1/RSK1, RPS6KA3/RSK2, RPS6KA2/RSK3, RPS6KA6/RSK4, SYK, MKNK1/MNK1, MKNK2/MNK2, RPS6KA5/MSK1, RPS6KA4/MSK2, MAPKAPK3 or MAPKAPK5) and phosphatases (such as DUSP1, DUSP4, DUSP6 or DUSP16) are other substrates which enable the propagation the MAPK/ERK signal to additional cytosolic and nuclear targets, thereby extending the specificity of the cascade.
- Gene Name:
- MAPK3
- Uniprot ID:
- P27361
- Molecular Weight:
- 43135.16 Da
References
- Kim G, Yurkow EJ: Chromium induces a persistent activation of mitogen-activated protein kinases by a redox-sensitive mechanism in H4 rat hepatoma cells. Cancer Res. 1996 May 1;56(9):2045-51. [8616849 ]
- General Function:
- Phospholipid-hydroperoxide glutathione peroxidase activity
- Specific Function:
- Protects cells against membrane lipid peroxidation and cell death. Required for normal sperm development and male fertility. Could play a major role in protecting mammals from the toxicity of ingested lipid hydroperoxides. Essential for embryonic development. Protects from radiation and oxidative damage.
- Gene Name:
- GPX4
- Uniprot ID:
- P36969
- Molecular Weight:
- 22174.52 Da
References
- Dillard CJ, Tappel AL: Mercury, silver, and gold inhibition of selenium-accelerated cysteine oxidation. J Inorg Biochem. 1986 Sep;28(1):13-20. [3760861 ]
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A1
- Uniprot ID:
- P05023
- Molecular Weight:
- 112895.01 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A2
- Uniprot ID:
- P50993
- Molecular Weight:
- 112264.385 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A3
- Uniprot ID:
- P13637
- Molecular Weight:
- 111747.51 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients. Plays a role in sperm motility.
- Gene Name:
- ATP1A4
- Uniprot ID:
- Q13733
- Molecular Weight:
- 114165.44 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The beta subunit regulates, through assembly of alpha/beta heterodimers, the number of sodium pumps transported to the plasma membrane.Involved in cell adhesion and establishing epithelial cell polarity.
- Gene Name:
- ATP1B1
- Uniprot ID:
- P05026
- Molecular Weight:
- 35061.07 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The exact function of the beta-2 subunit is not known.Mediates cell adhesion of neurons and astrocytes, and promotes neurite outgrowth.
- Gene Name:
- ATP1B2
- Uniprot ID:
- P14415
- Molecular Weight:
- 33366.925 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The exact function of the beta-3 subunit is not known.
- Gene Name:
- ATP1B3
- Uniprot ID:
- P54709
- Molecular Weight:
- 31512.34 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]
- General Function:
- Transporter activity
- Specific Function:
- May be involved in forming the receptor site for cardiac glycoside binding or may modulate the transport function of the sodium ATPase.
- Gene Name:
- FXYD2
- Uniprot ID:
- P54710
- Molecular Weight:
- 7283.265 Da
References
- Bianchini A, Playle RC, Wood CM, Walsh PJ: Mechanism of acute silver toxicity in marine invertebrates. Aquat Toxicol. 2005 Mar 25;72(1-2):67-82. Epub 2004 Dec 29. [15748748 ]