Trichloroethylene (T3D0016)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-03-06 18:57:55 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:20:52 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D0016 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Trichloroethylene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Trichloroethylene is a solvent and extractive in the manufacture of foods. One recent review of the epidemiology of kidney cancer rated cigarette smoking and obesity as more important risk factors for kidney cancer than exposure to solvents such as trichloroethylene. In contrast, the most recent overall assessment of human health risks associated with trichloroethylene states, [t]here is concordance between animal and human studies, which supports the conclusion that trichloroethylene is a potential kidney carcinogen. The evidence appears to be less certain at this time regarding the relationship between humans and liver cancer observed in mice, with the NAS suggesting that low-level exposure might not represent a significant liver cancer risk in the general population. The chemical compound trichloroethylene is a chlorinated hydrocarbon commonly used as an industrial solvent. It is a clear non-flammable liquid with a sweet smell. The first known report of TCE in groundwater was given in 1949 by two English public chemists who described two separate instances of well contamination by industrial releases of TCE. Based on available federal and state surveys, between 9% to 34% of the drinking water supply sources tested in the U.S. may have some TCE contamination, though EPA has reported that most water supplies are in compliance with the Maximum Contaminant Level (MCL) of 5 ppb. In addition, a growing concern in recent years at sites with TCE contamination in soil or groundwater has been vapor intrusion in buildings, which has resulted in indoor air exposures, such is in a recent case in the McCook Field Neighborhood of Dayton, Ohio. Trichloroethylene has been detected in 852 Superfund sites across the United States, according to the Agency for Toxic Substances and Disease Registry (ATSDR). Under the Safe Drinking Water Act of 1974, and as amended annual water quality testing is required for all public drinking water distributors. The EPA'S current guidelines for TCE can be found here. It should be noted that the EPA's table of TCE Releases to Ground is dated 1987 to 1993, thereby omitting one of the largest Superfund Cleanup sites in the nation, the NIBW in Scottsdale, Arizona. The TCE released here occurred prior to its appearance in the municipal drinking wells in 1982. This reaction can be catalyzed by a variety of substances. The most commonly used catalyst is a mixture of potassium chloride and aluminum chloride. However, various forms of porous carbon can also be used. This reaction produces tetrachloroethylene as a byproduct, and depending on the amount of chlorine fed to the reaction, tetrachloroethylene can even be the major product. Typically, trichloroethylene and tetrachloroethylene are collected together and then separated by distillation.Trichloroethylene: Parkinsonism and complex 1 mitochondrial neurotoxicity). Trichloroethylene is an effective solvent for a variety of organic materials. Trichloroethylene belongs to the family of Organochlorides. These are organic compounds containing a chlorine atom. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
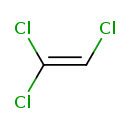
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C2HCl3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 131.388 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 129.914 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 1979-01-06 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 1,1,2-trichloroethene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | trichloroethylene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | ClC=C(Cl)Cl | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C2HCl3/c3-1-2(4)5/h1H | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=XSTXAVWGXDQKEL-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as vinyl chlorides. These are vinyl halides in which a chlorine atom is bonded to an sp2-hybridised carbon atom. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organohalogen compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Vinyl halides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Vinyl chlorides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Vinyl chlorides | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Liquid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Colorless liquid. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Oral (22) ; inhalation (22) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | The toxic and carcinogenic effects of trichloroethylene are believed to be caused mainly by its metabolites, including trichloroacetic acid, dichloroacetic acid, and chloral hydrate. The nephrotoxicity and nephrocarcinogenicity of TRI have been attributed to glutathione conjunction, which forms reactive, sulfur-containing metabolites. Dichloroacetic acid is known to inhibit pyruvate dehydrogenase kinase, while chloral hydrate inhibits alcohol dehydrogenase. Studies in rodents have shown that neurotoxic effects may be caused by trichloroethylene's incorporation into brain membranes or ability to alter the fatty acid pattern of brain phospholipids and amino acids. One of the mechanisms of trichloroethylene's carcinogenicity is believed to be the peroxisome proliferation induced by its metabolites. (22, 19, 1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Trichloroethylene is absorbed into the bloodstream and rapidly distributed throughout the body. Some is metabolized via cytochrome P-450 enzymes and the glutathione-conjugation pathway into metabolites such as trichloroacetic acid and trichloroethanol, which are excreted primarily in the urine. However, most trichoroethylene is exhaled unchanged or as carbon dioxide. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 2402 mg/kg (Oral, Mouse) (20) LD50: 20 001 mg/kg (Dermal, Rabbit) (20) LD50: 3222 mg/kg (Intraperitoneal, Mouse) (20) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | 3 to 5 mg/kg for an adult human. (26) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | 1, carcinogenic to humans. (25) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Trichloroethylene is mainly used as an industrial solvent, particularily to remove grease from metal parts. It is also an ingredient in adhesives, paint removers, typewriter correction fluids, and spot removers. Exposure may result from contact with contaminated water. (22) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Acute Inhalation: 2 ppm (24) Intermediate Inhalation: 0.1 ppm (24) Acute Oral: 0.2 mg/kg/day (24) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Chronic inhalation or ingestion of tricholoethylene causes nerve, kidney, and liver damage, impaired immune system function, impaired fetal development in pregnant women, and possibly death. It has also been linked to both kidney and liver cancer. (22) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Inhalation of trichloroethylene causes headaches, lung irritation, dizziness, poor coordination, and difficulty concentrating, while ingestion results in nausea. Larger amounts may cause unconciousness and impaired heart function. Skin contact often results in rashes. (22) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | There is no known antidote for trichloroethylene. Exposure is usually handled with symptomatic treatment. (23) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB29593 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 6575 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 13837280 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C06790 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 16602 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | TRICHLOROETHENE | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | D014241 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Trichloroethylene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | 1414 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Trichloroethylene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1.
- Gene Name:
- ESR1
- Uniprot ID:
- P03372
- Molecular Weight:
- 66215.45 Da
References
- Taccone-Gallucci M, Manca-di-Villahermosa S, Battistini L, Stuffler RG, Tedesco M, Maccarrone M: N-3 PUFAs reduce oxidative stress in ESRD patients on maintenance HD by inhibiting 5-lipoxygenase activity. Kidney Int. 2006 Apr;69(8):1450-4. [16531984 ]
- Luft S, Milki E, Glustrom E, Ampiah-Bonney R, O'Hara P. Binding of Organochloride and Pyrethroid Pesticides To Estrogen Receptors α and β: A Fluorescence Polarization Assay. Biophysical Journal 2009;96(3):444a.
- General Function:
- Zinc ion binding
- Specific Function:
- Nuclear hormone receptor. Binds estrogens with an affinity similar to that of ESR1, and activates expression of reporter genes containing estrogen response elements (ERE) in an estrogen-dependent manner (PubMed:20074560). Isoform beta-cx lacks ligand binding ability and has no or only very low ere binding activity resulting in the loss of ligand-dependent transactivation ability. DNA-binding by ESR1 and ESR2 is rapidly lost at 37 degrees Celsius in the absence of ligand while in the presence of 17 beta-estradiol and 4-hydroxy-tamoxifen loss in DNA-binding at elevated temperature is more gradual.
- Gene Name:
- ESR2
- Uniprot ID:
- Q92731
- Molecular Weight:
- 59215.765 Da
References
- Taccone-Gallucci M, Manca-di-Villahermosa S, Battistini L, Stuffler RG, Tedesco M, Maccarrone M: N-3 PUFAs reduce oxidative stress in ESRD patients on maintenance HD by inhibiting 5-lipoxygenase activity. Kidney Int. 2006 Apr;69(8):1450-4. [16531984 ]
- Luft S, Milki E, Glustrom E, Ampiah-Bonney R, O'Hara P. Binding of Organochloride and Pyrethroid Pesticides To Estrogen Receptors α and β: A Fluorescence Polarization Assay. Biophysical Journal 2009;96(3):444a.
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1A
- Uniprot ID:
- P07327
- Molecular Weight:
- 39858.37 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1B
- Uniprot ID:
- P00325
- Molecular Weight:
- 39854.21 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH1C
- Uniprot ID:
- P00326
- Molecular Weight:
- 39867.27 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH4
- Uniprot ID:
- P08319
- Molecular Weight:
- 40221.335 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Not Available
- Gene Name:
- ADH6
- Uniprot ID:
- P28332
- Molecular Weight:
- 39088.335 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Could function in retinol oxidation for the synthesis of retinoic acid, a hormone important for cellular differentiation. Medium-chain (octanol) and aromatic (m-nitrobenzaldehyde) compounds are the best substrates. Ethanol is not a good substrate but at the high ethanol concentrations reached in the digestive tract, it plays a role in the ethanol oxidation and contributes to the first pass ethanol metabolism.
- Gene Name:
- ADH7
- Uniprot ID:
- P40394
- Molecular Weight:
- 41480.985 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Class-III ADH is remarkably ineffective in oxidizing ethanol, but it readily catalyzes the oxidation of long-chain primary alcohols and the oxidation of S-(hydroxymethyl) glutathione.
- Gene Name:
- ADH5
- Uniprot ID:
- P11766
- Molecular Weight:
- 39723.945 Da
References
- Sharkawi M, De Saint Blanquat G, Elfassy B: Inhibition of alcohol dehydrogenase by chloral hydrate and trichloroethanol: possible role in the chloral hydrate-ethanol interaction. Toxicol Lett. 1983 Jul;17(3-4):321-8. [6353674 ]
- General Function:
- Signal transducer activity
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of the calcium.
- Gene Name:
- ATP2C1
- Uniprot ID:
- P98194
- Molecular Weight:
- 100576.42 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Metal ion binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium.
- Gene Name:
- ATP2C2
- Uniprot ID:
- O75185
- Molecular Weight:
- 103186.475 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel (By similarity).
- Gene Name:
- GABRA1
- Uniprot ID:
- P14867
- Molecular Weight:
- 51801.395 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRA2
- Uniprot ID:
- P47869
- Molecular Weight:
- 51325.85 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRA3
- Uniprot ID:
- P34903
- Molecular Weight:
- 55164.055 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRA4
- Uniprot ID:
- P48169
- Molecular Weight:
- 61622.645 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Transporter activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRA5
- Uniprot ID:
- P31644
- Molecular Weight:
- 52145.645 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRA6
- Uniprot ID:
- Q16445
- Molecular Weight:
- 51023.69 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel (By similarity).
- Gene Name:
- GABRB1
- Uniprot ID:
- P18505
- Molecular Weight:
- 54234.085 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel.
- Gene Name:
- GABRB2
- Uniprot ID:
- P47870
- Molecular Weight:
- 59149.895 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-gated chloride ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel.
- Gene Name:
- GABRB3
- Uniprot ID:
- P28472
- Molecular Weight:
- 54115.04 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-a receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRD
- Uniprot ID:
- O14764
- Molecular Weight:
- 50707.835 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRE
- Uniprot ID:
- P78334
- Molecular Weight:
- 57971.175 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRG1
- Uniprot ID:
- Q8N1C3
- Molecular Weight:
- 53594.49 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- Component of the heteropentameric receptor for GABA, the major inhibitory neurotransmitter in the vertebrate brain. Functions also as histamine receptor and mediates cellular responses to histamine. Functions as receptor for diazepines and various anesthetics, such as pentobarbital; these are bound at a separate allosteric effector binding site. Functions as ligand-gated chloride channel.
- Gene Name:
- GABRG2
- Uniprot ID:
- P18507
- Molecular Weight:
- 54161.78 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Inhibitory extracellular ligand-gated ion channel activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRG3
- Uniprot ID:
- Q99928
- Molecular Weight:
- 54288.16 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-a receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel. In the uterus, the function of the receptor appears to be related to tissue contractility. The binding of this pI subunit with other GABA(A) receptor subunits alters the sensitivity of recombinant receptors to modulatory agents such as pregnanolone.
- Gene Name:
- GABRP
- Uniprot ID:
- O00591
- Molecular Weight:
- 50639.735 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-a receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel. Rho-1 GABA receptor could play a role in retinal neurotransmission.
- Gene Name:
- GABRR1
- Uniprot ID:
- P24046
- Molecular Weight:
- 55882.91 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-a receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel. Rho-2 GABA receptor could play a role in retinal neurotransmission.
- Gene Name:
- GABRR2
- Uniprot ID:
- P28476
- Molecular Weight:
- 54150.41 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Gaba-a receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRR3
- Uniprot ID:
- A8MPY1
- Molecular Weight:
- 54271.1 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- GABA, the major inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel.
- Gene Name:
- GABRQ
- Uniprot ID:
- Q9UN88
- Molecular Weight:
- 72020.875 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Purinergic nucleotide receptor activity
- Specific Function:
- Receptor for ATP that acts as a ligand-gated ion channel.
- Gene Name:
- P2RX3
- Uniprot ID:
- P56373
- Molecular Weight:
- 44288.65 Da
References
- Fischer W, Wirkner K, Weber M, Eberts C, Koles L, Reinhardt R, Franke H, Allgaier C, Gillen C, Illes P: Characterization of P2X3, P2Y1 and P2Y4 receptors in cultured HEK293-hP2X3 cells and their inhibition by ethanol and trichloroethanol. J Neurochem. 2003 May;85(3):779-90. [12694404 ]
- General Function:
- Pdz domain binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell.
- Gene Name:
- ATP2B1
- Uniprot ID:
- P20020
- Molecular Weight:
- 138754.045 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Protein c-terminus binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell.
- Gene Name:
- ATP2B2
- Uniprot ID:
- Q01814
- Molecular Weight:
- 136875.18 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Pdz domain binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell.
- Gene Name:
- ATP2B3
- Uniprot ID:
- Q16720
- Molecular Weight:
- 134196.025 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Scaffold protein binding
- Specific Function:
- Calcium/calmodulin-regulated and magnesium-dependent enzyme that catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell (PubMed:8530416). By regulating sperm cell calcium homeostasis, may play a role in sperm motility (By similarity).
- Gene Name:
- ATP2B4
- Uniprot ID:
- P23634
- Molecular Weight:
- 137919.03 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Protein homodimerization activity
- Specific Function:
- Key regulator of striated muscle performance by acting as the major Ca(2+) ATPase responsible for the reuptake of cytosolic Ca(2+) into the sarcoplasmic reticulum. Catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen. Contributes to calcium sequestration involved in muscular excitation/contraction.
- Gene Name:
- ATP2A1
- Uniprot ID:
- O14983
- Molecular Weight:
- 110251.36 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- S100 protein binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen. Isoform 2 is involved in the regulation of the contraction/relaxation cycle.
- Gene Name:
- ATP2A2
- Uniprot ID:
- P16615
- Molecular Weight:
- 114755.765 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Metal ion binding
- Specific Function:
- This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium. Transports calcium ions from the cytosol into the sarcoplasmic/endoplasmic reticulum lumen. Contributes to calcium sequestration involved in muscular excitation/contraction.
- Gene Name:
- ATP2A3
- Uniprot ID:
- Q93084
- Molecular Weight:
- 113976.23 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A1
- Uniprot ID:
- P05023
- Molecular Weight:
- 112895.01 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A2
- Uniprot ID:
- P50993
- Molecular Weight:
- 112264.385 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Steroid hormone binding
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
- Gene Name:
- ATP1A3
- Uniprot ID:
- P13637
- Molecular Weight:
- 111747.51 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients. Plays a role in sperm motility.
- Gene Name:
- ATP1A4
- Uniprot ID:
- Q13733
- Molecular Weight:
- 114165.44 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The beta subunit regulates, through assembly of alpha/beta heterodimers, the number of sodium pumps transported to the plasma membrane.Involved in cell adhesion and establishing epithelial cell polarity.
- Gene Name:
- ATP1B1
- Uniprot ID:
- P05026
- Molecular Weight:
- 35061.07 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The exact function of the beta-2 subunit is not known.Mediates cell adhesion of neurons and astrocytes, and promotes neurite outgrowth.
- Gene Name:
- ATP1B2
- Uniprot ID:
- P14415
- Molecular Weight:
- 33366.925 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Sodium:potassium-exchanging atpase activity
- Specific Function:
- This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The exact function of the beta-3 subunit is not known.
- Gene Name:
- ATP1B3
- Uniprot ID:
- P54709
- Molecular Weight:
- 31512.34 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Transporter activity
- Specific Function:
- May be involved in forming the receptor site for cardiac glycoside binding or may modulate the transport function of the sodium ATPase.
- Gene Name:
- FXYD2
- Uniprot ID:
- P54710
- Molecular Weight:
- 7283.265 Da
References
- Casarett LJ, Klaassen CD, and Watkins JB (2003). Casarett and Doull's essentials of toxicology. New York: McGraw-Hill/Medical Pub. Div.
- General Function:
- Pyruvate dehydrogenase (acetyl-transferring) kinase activity
- Specific Function:
- Kinase that plays a key role in regulation of glucose and fatty acid metabolism and homeostasis via phosphorylation of the pyruvate dehydrogenase subunits PDHA1 and PDHA2. This inhibits pyruvate dehydrogenase activity, and thereby regulates metabolite flux through the tricarboxylic acid cycle, down-regulates aerobic respiration and inhibits the formation of acetyl-coenzyme A from pyruvate. Plays an important role in cellular responses to hypoxia and is important for cell proliferation under hypoxia. Protects cells against apoptosis in response to hypoxia and oxidative stress.
- Gene Name:
- PDK1
- Uniprot ID:
- Q15118
- Molecular Weight:
- 49243.765 Da
References
- Lantum HB, Baggs RB, Krenitsky DM, Anders MW: Nephrotoxicity of chlorofluoroacetic acid in rats. Toxicol Sci. 2002 Dec;70(2):261-8. [12441371 ]
- General Function:
- Pyruvate dehydrogenase (acetyl-transferring) kinase activity
- Specific Function:
- Kinase that plays a key role in the regulation of glucose and fatty acid metabolism and homeostasis via phosphorylation of the pyruvate dehydrogenase subunits PDHA1 and PDHA2. This inhibits pyruvate dehydrogenase activity, and thereby regulates metabolite flux through the tricarboxylic acid cycle, down-regulates aerobic respiration and inhibits the formation of acetyl-coenzyme A from pyruvate. Inhibition of pyruvate dehydrogenase decreases glucose utilization and increases fat metabolism. Mediates cellular responses to insulin. Plays an important role in maintaining normal blood glucose levels and in metabolic adaptation to nutrient availability. Via its regulation of pyruvate dehydrogenase activity, plays an important role in maintaining normal blood pH and in preventing the accumulation of ketone bodies under starvation. Plays a role in the regulation of cell proliferation and in resistance to apoptosis under oxidative stress. Plays a role in p53/TP53-mediated apoptosis.
- Gene Name:
- PDK2
- Uniprot ID:
- Q15119
- Molecular Weight:
- 46153.39 Da
References
- Lantum HB, Baggs RB, Krenitsky DM, Anders MW: Nephrotoxicity of chlorofluoroacetic acid in rats. Toxicol Sci. 2002 Dec;70(2):261-8. [12441371 ]
- General Function:
- Pyruvate dehydrogenase (acetyl-transferring) kinase activity
- Specific Function:
- Inhibits pyruvate dehydrogenase activity by phosphorylation of the E1 subunit PDHA1, and thereby regulates glucose metabolism and aerobic respiration. Can also phosphorylate PDHA2. Decreases glucose utilization and increases fat metabolism in response to prolonged fasting, and as adaptation to a high-fat diet. Plays a role in glucose homeostasis and in maintaining normal blood glucose levels in function of nutrient levels and under starvation. Plays a role in the generation of reactive oxygen species.
- Gene Name:
- PDK3
- Uniprot ID:
- Q15120
- Molecular Weight:
- 46938.485 Da
References
- Lantum HB, Baggs RB, Krenitsky DM, Anders MW: Nephrotoxicity of chlorofluoroacetic acid in rats. Toxicol Sci. 2002 Dec;70(2):261-8. [12441371 ]
- General Function:
- Pyruvate dehydrogenase (acetyl-transferring) kinase activity
- Specific Function:
- Kinase that plays a key role in regulation of glucose and fatty acid metabolism and homeostasis via phosphorylation of the pyruvate dehydrogenase subunits PDHA1 and PDHA2. This inhibits pyruvate dehydrogenase activity, and thereby regulates metabolite flux through the tricarboxylic acid cycle, down-regulates aerobic respiration and inhibits the formation of acetyl-coenzyme A from pyruvate. Inhibition of pyruvate dehydrogenase decreases glucose utilization and increases fat metabolism in response to prolonged fasting and starvation. Plays an important role in maintaining normal blood glucose levels under starvation, and is involved in the insulin signaling cascade. Via its regulation of pyruvate dehydrogenase activity, plays an important role in maintaining normal blood pH and in preventing the accumulation of ketone bodies under starvation. In the fed state, mediates cellular responses to glucose levels and to a high-fat diet. Regulates both fatty acid oxidation and de novo fatty acid biosynthesis. Plays a role in the generation of reactive oxygen species. Protects detached epithelial cells against anoikis. Plays a role in cell proliferation via its role in regulating carbohydrate and fatty acid metabolism.
- Gene Name:
- PDK4
- Uniprot ID:
- Q16654
- Molecular Weight:
- 46468.79 Da
References
- Lantum HB, Baggs RB, Krenitsky DM, Anders MW: Nephrotoxicity of chlorofluoroacetic acid in rats. Toxicol Sci. 2002 Dec;70(2):261-8. [12441371 ]