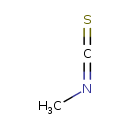Methyl isothiocyanate (T3D0780)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2009-05-25 19:43:14 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2014-12-24 20:22:49 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession Number | T3D0780 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Methyl isothiocyanate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Small Molecule | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Methyl isothiocyanate is found in horseradish. Methyl isothiocyanate is isolated from mustard oil, present as glucoside (Methyl glucosinolate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Compound Type |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C2H3NS | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Mass | 73.117 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Mass | 72.999 g/mol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 556-61-6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | isothiocyanatomethane | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | methyl isothiocyanate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | CN=C=S | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C2H3NS/c1-3-2-4/h1H3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | InChIKey=LGDSHSYDSCRFAB-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as isothiocyanates. These are organic compounds containing the isothiocyanate group, an isocyanate analogue with the general formula RN=C=S. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organosulfur compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Isothiocyanates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Isothiocyanates | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin | Exogenous | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofluid Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Tissue Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Applications | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Roles | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Appearance | Colorless crystals. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Profile | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Route of Exposure | Inhalation (5) ; oral (5) ; dermal (5) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mechanism of Toxicity | Cyanide is an inhibitor of cytochrome c oxidase in the fourth complex of the electron transport chain (found in the membrane of the mitochondria of eukaryotic cells). It complexes with the ferric iron atom in this enzyme. The binding of cyanide to this cytochrome prevents transport of electrons from cytochrome c oxidase to oxygen. As a result, the electron transport chain is disrupted and the cell can no longer aerobically produce ATP for energy. Tissues that mainly depend on aerobic respiration, such as the central nervous system and the heart, are particularly affected. Cyanide is also known produce some of its toxic effects by binding to catalase, glutathione peroxidase, methemoglobin, hydroxocobalamin, phosphatase, tyrosinase, ascorbic acid oxidase, xanthine oxidase, succinic dehydrogenase, and Cu/Zn superoxide dismutase. Cyanide binds to the ferric ion of methemoglobin to form inactive cyanmethemoglobin. (6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolism | Cyanide is rapidly alsorbed through oral, inhalation, and dermal routes and distributed throughout the body. Cyanide is mainly metabolized into thiocyanate by either rhodanese or 3-mercaptopyruvate sulfur transferase. Cyanide metabolites are excreted in the urine. (5) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Toxicity Values | LD50: 90-104 mg/kg (Oral, Mouse) (2) LD50: 54 mg/kg (Intraperitoneal, Rat) (1) LD50: 59 mg/kg (Subcutaneous, Rat) (1) LD50: 2780 mg/kg (Dermal, Rat) (1) LC50: 1900 mg/m3 over 1 hour (Inhalation, Rat) (1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Lethal Dose | 200 to 300 milligrams for an adult human (cyanide salts). (3) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Carcinogenicity (IARC Classification) | No indication of carcinogenicity to humans (not listed by IARC). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Uses/Sources | Methyl isothiocyanate is a precursor to a variety of valuable bioactive compounds and is also used in agriculture as a soil fumigant. (7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Minimum Risk Level | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Health Effects | Exposure to high levels of cyanide for a short time harms the brain and heart and can even cause coma, seizures, apnea, cardiac arrest and death. Chronic inhalation of cyanide causes breathing difficulties, chest pain, vomiting, blood changes, headaches, and enlargement of the thyroid gland. Skin contact with cyanide salts can irritate and produce sores. (5, 6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | Cyanide poisoning is identified by rapid, deep breathing and shortness of breath, general weakness, giddiness, headaches, vertigo, confusion, convulsions/seizures and eventually loss of consciousness. (5, 6) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Treatment | EYES: irrigate opened eyes for several minutes under running water. INGESTION: do not induce vomiting. Rinse mouth with water (never give anything by mouth to an unconscious person). Seek immediate medical advice. SKIN: should be treated immediately by rinsing the affected parts in cold running water for at least 15 minutes, followed by thorough washing with soap and water. If necessary, the person should shower and change contaminated clothing and shoes, and then must seek medical attention. INHALATION: supply fresh air. If required provide artificial respiration. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB34106 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound ID | 11167 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEMBL ID | CHEMBL396000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChemSpider ID | 10694 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG ID | C18587 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UniProt ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| OMIM ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 78337 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | CPD-10560 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CTD ID | C005227 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stitch ID | Methyl isothiocyanate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ACToR ID | 3781 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MSDS | Link | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Regulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Up-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Down-Regulated Genes | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Targets
- General Function:
- Metal ion binding
- Specific Function:
- Not Available
- Gene Name:
- ALPPL2
- Uniprot ID:
- P10696
- Molecular Weight:
- 57376.515 Da
References
- Gerbitz KD: Human alkaline phosphatases. II. Metalloenzyme properties of the enzyme from human liver. Hoppe Seylers Z Physiol Chem. 1977 Nov;358(11):1491-7. [924371 ]
- General Function:
- Pyrophosphatase activity
- Specific Function:
- This isozyme may play a role in skeletal mineralization.
- Gene Name:
- ALPL
- Uniprot ID:
- P05186
- Molecular Weight:
- 57304.435 Da
References
- Gerbitz KD: Human alkaline phosphatases. II. Metalloenzyme properties of the enzyme from human liver. Hoppe Seylers Z Physiol Chem. 1977 Nov;358(11):1491-7. [924371 ]
- General Function:
- Receptor binding
- Specific Function:
- Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide. Promotes growth of cells including T-cells, B-cells, myeloid leukemia cells, melanoma cells, mastocytoma cells and normal and transformed fibroblast cells.
- Gene Name:
- CAT
- Uniprot ID:
- P04040
- Molecular Weight:
- 59755.82 Da
References
- Kang YS, Lee DH, Yoon BJ, Oh DC: Purification and characterization of a catalase from photosynthetic bacterium Rhodospirillum rubrum S1 grown under anaerobic conditions. J Microbiol. 2006 Apr;44(2):185-91. [16728955 ]
- General Function:
- Iron ion binding
- Specific Function:
- Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Subunits 1-3 form the functional core of the enzyme complex. CO I is the catalytic subunit of the enzyme. Electrons originating in cytochrome c are transferred via the copper A center of subunit 2 and heme A of subunit 1 to the bimetallic center formed by heme A3 and copper B.
- Gene Name:
- MT-CO1
- Uniprot ID:
- P00395
- Molecular Weight:
- 57040.91 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Subunits 1-3 form the functional core of the enzyme complex. Subunit 2 transfers the electrons from cytochrome c via its binuclear copper A center to the bimetallic center of the catalytic subunit 1.
- Gene Name:
- MT-CO2
- Uniprot ID:
- P00403
- Molecular Weight:
- 25564.73 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX4I1
- Uniprot ID:
- P13073
- Molecular Weight:
- 19576.6 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX4I2
- Uniprot ID:
- Q96KJ9
- Molecular Weight:
- 20010.02 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Metal ion binding
- Specific Function:
- This is the heme A-containing chain of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX5A
- Uniprot ID:
- P20674
- Molecular Weight:
- 16761.985 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Metal ion binding
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX5B
- Uniprot ID:
- P10606
- Molecular Weight:
- 13695.57 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX6A1
- Uniprot ID:
- P12074
- Molecular Weight:
- 12154.8 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX6A2
- Uniprot ID:
- Q02221
- Molecular Weight:
- 10815.32 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX6C
- Uniprot ID:
- P09669
- Molecular Weight:
- 8781.36 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX7A1
- Uniprot ID:
- P24310
- Molecular Weight:
- 9117.44 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX7A2
- Uniprot ID:
- P14406
- Molecular Weight:
- 9395.89 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport. Plays a role in proper central nervous system (CNS) development in vertebrates.
- Gene Name:
- COX7B
- Uniprot ID:
- P24311
- Molecular Weight:
- 9160.485 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX7B2
- Uniprot ID:
- Q8TF08
- Molecular Weight:
- 9077.43 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX7C
- Uniprot ID:
- P15954
- Molecular Weight:
- 7245.45 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX8A
- Uniprot ID:
- P10176
- Molecular Weight:
- 7579.0 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Cytochrome-c oxidase activity
- Specific Function:
- This protein is one of the nuclear-coded polypeptide chains of cytochrome c oxidase, the terminal oxidase in mitochondrial electron transport.
- Gene Name:
- COX8C
- Uniprot ID:
- Q7Z4L0
- Molecular Weight:
- 8128.575 Da
References
- Wikipedia. Cyanide poisoning. Last Updated 30 March 2009. [Link]
- General Function:
- Glutathione peroxidase activity
- Specific Function:
- Protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide, by glutathione. May constitute a glutathione peroxidase-like protective system against peroxide damage in sperm membrane lipids.
- Gene Name:
- GPX5
- Uniprot ID:
- O75715
- Molecular Weight:
- 25202.14 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Protect the extracellular space from toxic effect of reactive oxygen intermediates by converting superoxide radicals into hydrogen peroxide and oxygen.
- Gene Name:
- SOD3
- Uniprot ID:
- P08294
- Molecular Weight:
- 25850.675 Da
References
- Lee WG, Hwang JH, Na BK, Cho JH, Lee HW, Cho SH, Kong Y, Song CY, Kim TS: Functional expression of a recombinant copper/zinc superoxide dismutase of filarial nematode, Brugia malayi. J Parasitol. 2005 Feb;91(1):205-8. [15856906 ]
- General Function:
- Sh3 domain binding
- Specific Function:
- Protects the hemoglobin in erythrocytes from oxidative breakdown.
- Gene Name:
- GPX1
- Uniprot ID:
- P07203
- Molecular Weight:
- 22087.94 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Glutathione peroxidase activity
- Specific Function:
- Could play a major role in protecting mammals from the toxicity of ingested organic hydroperoxides. Tert-butyl hydroperoxide, cumene hydroperoxide and linoleic acid hydroperoxide but not phosphatidycholine hydroperoxide, can act as acceptors.
- Gene Name:
- GPX2
- Uniprot ID:
- P18283
- Molecular Weight:
- 21953.835 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Transcription factor binding
- Specific Function:
- Protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide, by glutathione.
- Gene Name:
- GPX3
- Uniprot ID:
- P22352
- Molecular Weight:
- 25552.185 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Peroxidase activity
- Specific Function:
- It protects esophageal epithelia from hydrogen peroxide-induced oxidative stress. It suppresses acidic bile acid-induced reactive oxigen species (ROS) and protects against oxidative DNA damage and double-strand breaks.
- Gene Name:
- GPX7
- Uniprot ID:
- Q96SL4
- Molecular Weight:
- 20995.88 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Nadp binding
- Specific Function:
- Maintains high levels of reduced glutathione in the cytosol.
- Gene Name:
- GSR
- Uniprot ID:
- P00390
- Molecular Weight:
- 56256.565 Da
References
- Ardelt BK, Borowitz JL, Isom GE: Brain lipid peroxidation and antioxidant protectant mechanisms following acute cyanide intoxication. Toxicology. 1989 Jun 1;56(2):147-54. [2734799 ]
- General Function:
- Phospholipid-hydroperoxide glutathione peroxidase activity
- Specific Function:
- Protects cells against membrane lipid peroxidation and cell death. Required for normal sperm development and male fertility. Could play a major role in protecting mammals from the toxicity of ingested lipid hydroperoxides. Essential for embryonic development. Protects from radiation and oxidative damage.
- Gene Name:
- GPX4
- Uniprot ID:
- P36969
- Molecular Weight:
- 22174.52 Da
References
- Kraus RJ, Ganther HE: Reaction of cyanide with glutathione peroxidase. Biochem Biophys Res Commun. 1980 Oct 16;96(3):1116-22. [7437059 ]
- General Function:
- Ubiquinone binding
- Specific Function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
- Gene Name:
- SDHD
- Uniprot ID:
- O14521
- Molecular Weight:
- 17042.82 Da
References
- Ardelt BK, Borowitz JL, Isom GE: Brain lipid peroxidation and antioxidant protectant mechanisms following acute cyanide intoxication. Toxicology. 1989 Jun 1;56(2):147-54. [2734799 ]
- General Function:
- Succinate dehydrogenase activity
- Specific Function:
- Flavoprotein (FP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q). Can act as a tumor suppressor.
- Gene Name:
- SDHA
- Uniprot ID:
- P31040
- Molecular Weight:
- 72690.975 Da
References
- Ardelt BK, Borowitz JL, Isom GE: Brain lipid peroxidation and antioxidant protectant mechanisms following acute cyanide intoxication. Toxicology. 1989 Jun 1;56(2):147-54. [2734799 ]
- General Function:
- Ubiquinone binding
- Specific Function:
- Iron-sulfur protein (IP) subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
- Gene Name:
- SDHB
- Uniprot ID:
- P21912
- Molecular Weight:
- 31629.365 Da
References
- Ardelt BK, Borowitz JL, Isom GE: Brain lipid peroxidation and antioxidant protectant mechanisms following acute cyanide intoxication. Toxicology. 1989 Jun 1;56(2):147-54. [2734799 ]
- General Function:
- Succinate dehydrogenase activity
- Specific Function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
- Gene Name:
- SDHC
- Uniprot ID:
- Q99643
- Molecular Weight:
- 18610.03 Da
References
- Ardelt BK, Borowitz JL, Isom GE: Brain lipid peroxidation and antioxidant protectant mechanisms following acute cyanide intoxication. Toxicology. 1989 Jun 1;56(2):147-54. [2734799 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Destroys radicals which are normally produced within the cells and which are toxic to biological systems.
- Gene Name:
- SOD1
- Uniprot ID:
- P00441
- Molecular Weight:
- 15935.685 Da
References
- Lee WG, Hwang JH, Na BK, Cho JH, Lee HW, Cho SH, Kong Y, Song CY, Kim TS: Functional expression of a recombinant copper/zinc superoxide dismutase of filarial nematode, Brugia malayi. J Parasitol. 2005 Feb;91(1):205-8. [15856906 ]
- General Function:
- Protein homodimerization activity
- Specific Function:
- This is a copper-containing oxidase that functions in the formation of pigments such as melanins and other polyphenolic compounds. Catalyzes the rate-limiting conversions of tyrosine to DOPA, DOPA to DOPA-quinone and possibly 5,6-dihydroxyindole to indole-5,6 quinone.
- Gene Name:
- TYR
- Uniprot ID:
- P14679
- Molecular Weight:
- 60392.69 Da
References
- Laufer Z, Beckett RP, Minibayeva FV: Co-occurrence of the multicopper oxidases tyrosinase and laccase in lichens in sub-order peltigerineae. Ann Bot. 2006 Nov;98(5):1035-42. Epub 2006 Sep 1. [16950829 ]
- General Function:
- Scaffold protein binding
- Specific Function:
- Receptor for extracellular adenine nucleotides such as ATP and ADP. In platelets binding to ADP leads to mobilization of intracellular calcium ions via activation of phospholipase C, a change in platelet shape, and probably to platelet aggregation.
- Gene Name:
- P2RY1
- Uniprot ID:
- P47900
- Molecular Weight:
- 42071.08 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 8.10 uM | NVS_GPCR_hPY2 | Novascreen |
| AC50 | 0.63 uM | NVS_GPCR_hPY2 | Novascreen |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Transmembrane signaling receptor activity
- Specific Function:
- Involved in lymphocyte proliferation and functions as a signal transmitting receptor in lymphocytes, natural killer (NK) cells, and platelets.
- Gene Name:
- CD69
- Uniprot ID:
- Q07108
- Molecular Weight:
- 22559.25 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.44 uM | BSK_SAg_CD69_down | BioSeek |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Steroid hydroxylase activity
- Specific Function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics. Acts as a 1,4-cineole 2-exo-monooxygenase.
- Gene Name:
- CYP2B6
- Uniprot ID:
- P20813
- Molecular Weight:
- 56277.81 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 4.53 uM | CLZD_CYP2B6_6 | CellzDirect |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]
- General Function:
- Zinc ion binding
- Specific Function:
- Isoform Alpha-1: Nuclear hormone receptor that can act as a repressor or activator of transcription. High affinity receptor for thyroid hormones, including triiodothyronine and thyroxine.Isoform Alpha-2: Does not bind thyroid hormone and functions as a weak dominant negative inhibitor of thyroid hormone action.
- Gene Name:
- THRA
- Uniprot ID:
- P10827
- Molecular Weight:
- 54815.055 Da
Binding/Activity Constants
| Type | Value | Assay Type | Assay Source |
|---|---|---|---|
| AC50 | 9.90 uM | ATG_THRa1_TRANS | Attagene |
References
- Sipes NS, Martin MT, Kothiya P, Reif DM, Judson RS, Richard AM, Houck KA, Dix DJ, Kavlock RJ, Knudsen TB: Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem Res Toxicol. 2013 Jun 17;26(6):878-95. doi: 10.1021/tx400021f. Epub 2013 May 16. [23611293 ]